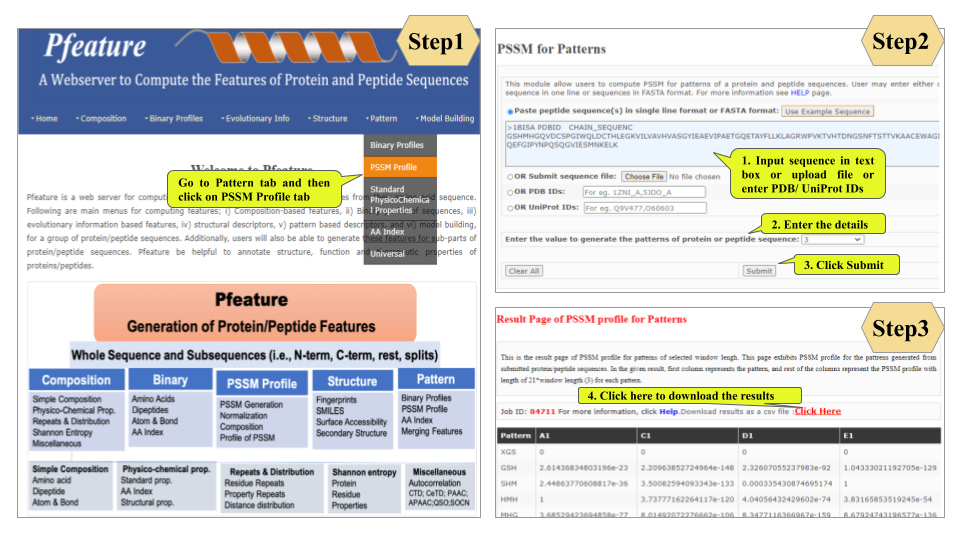
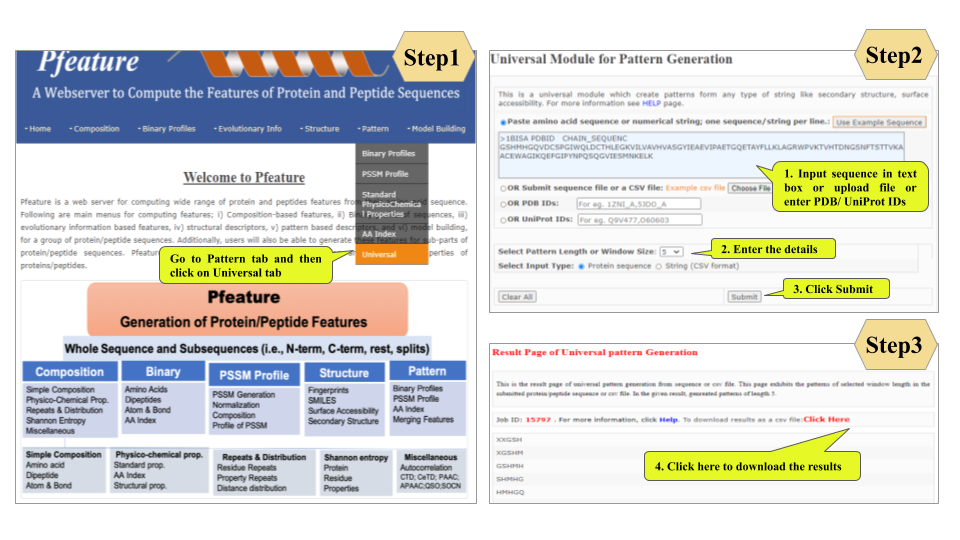
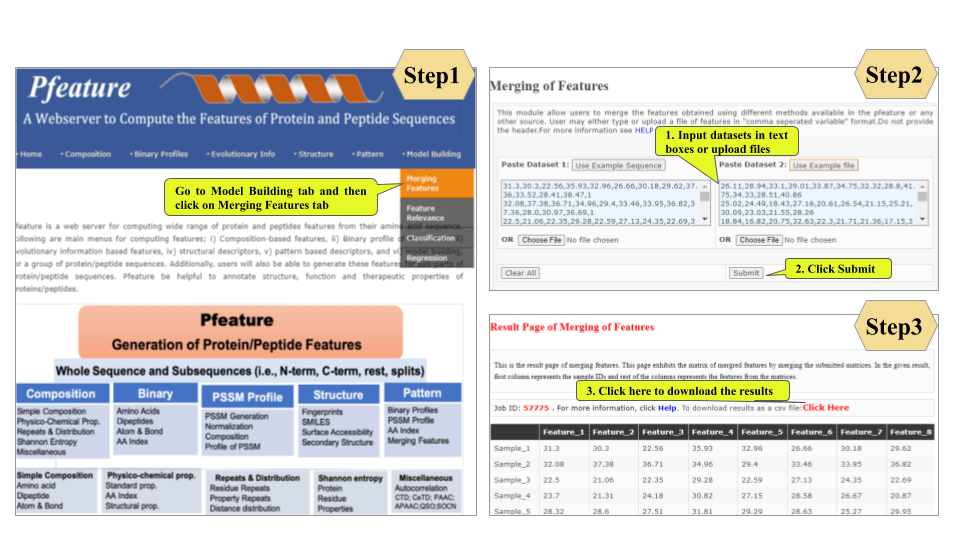
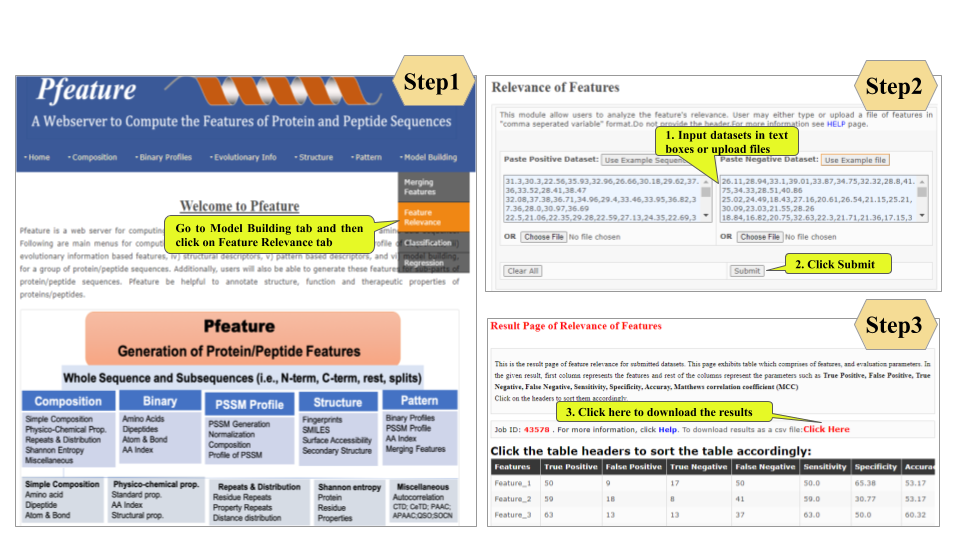
Welcome to Help Page
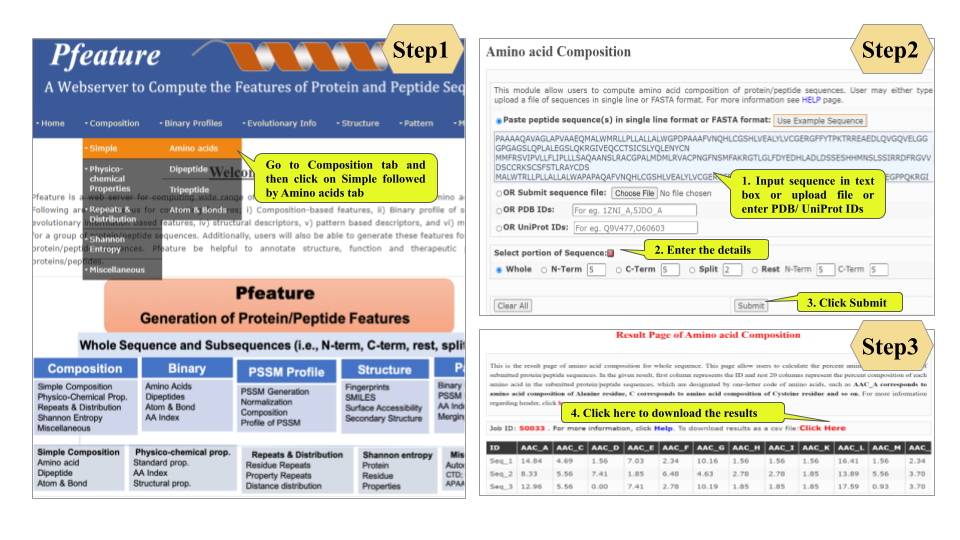
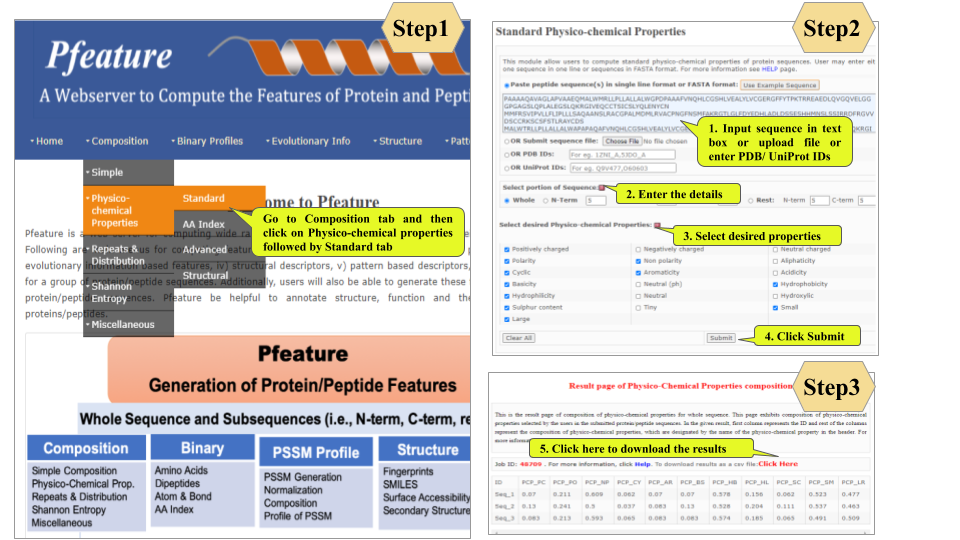
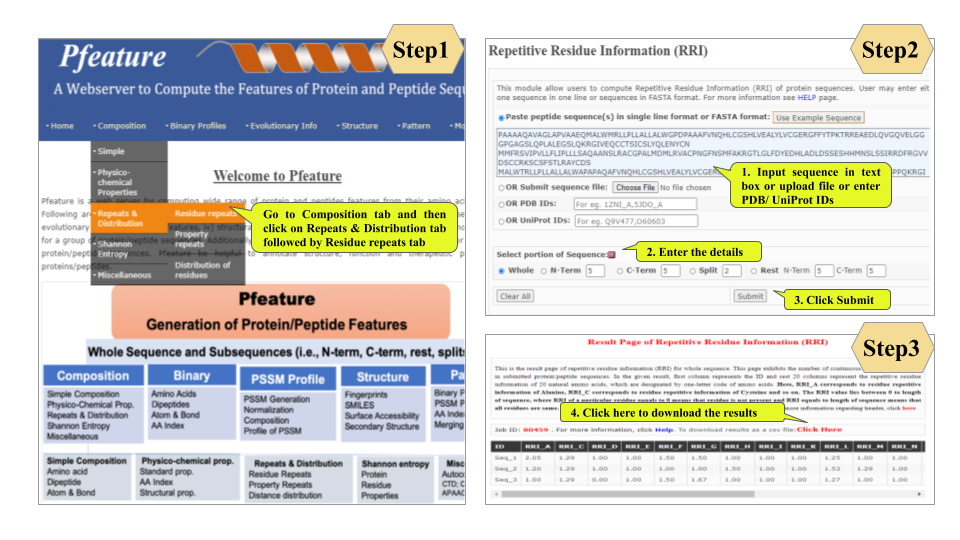
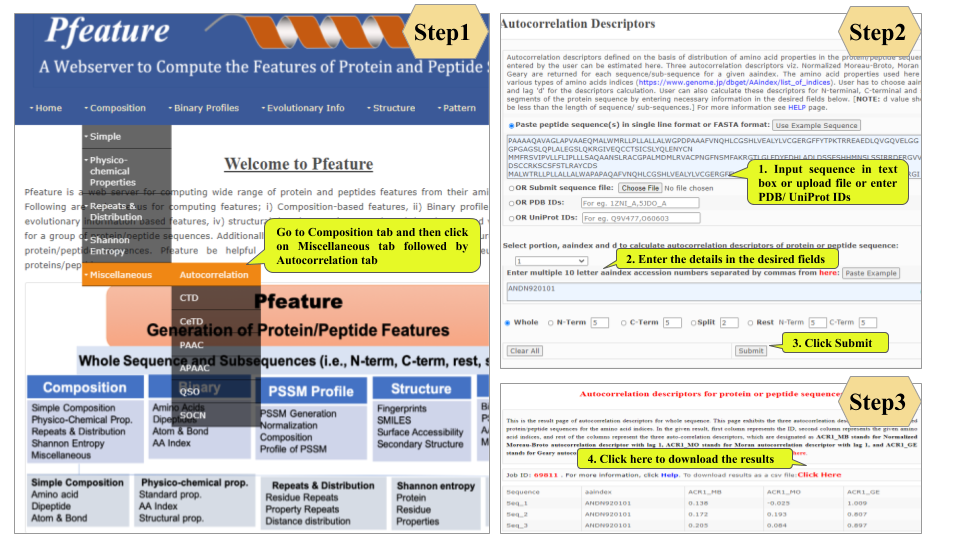
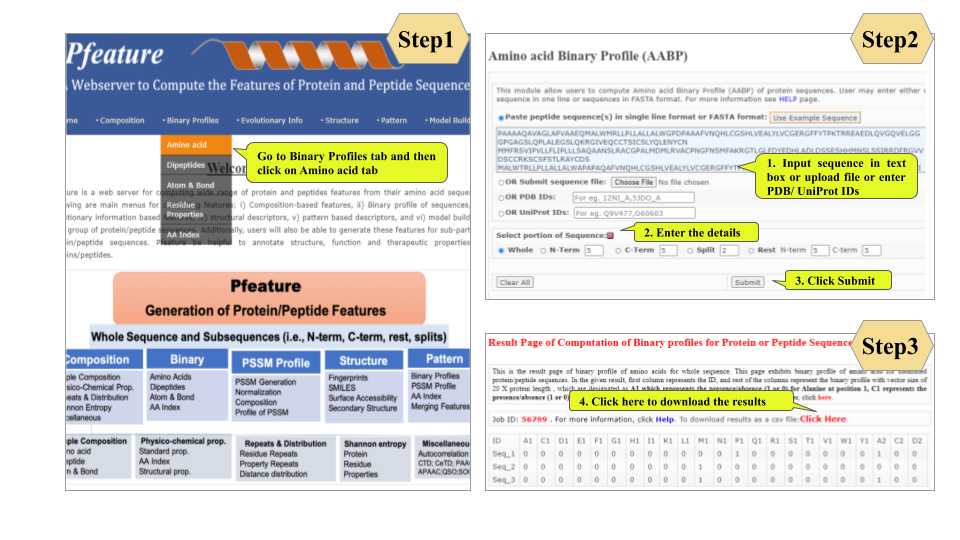
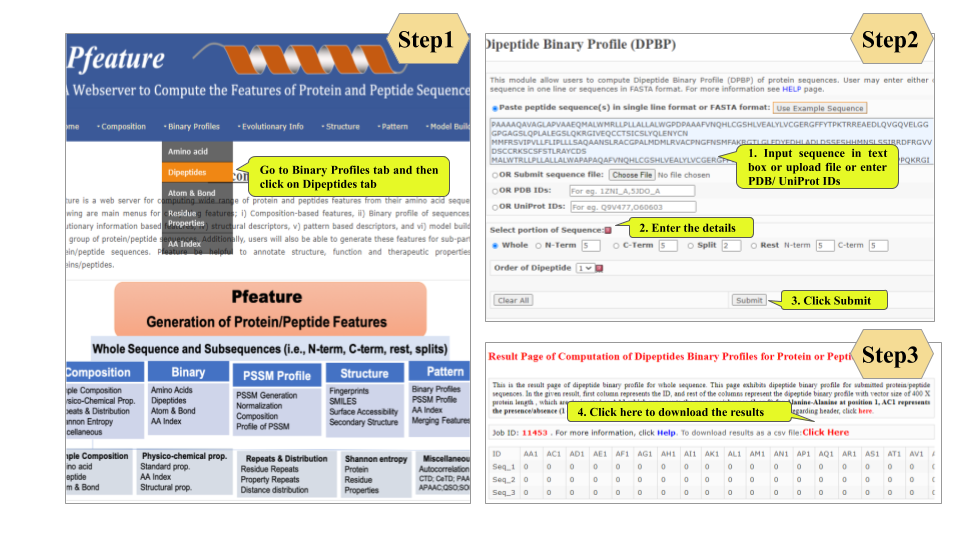
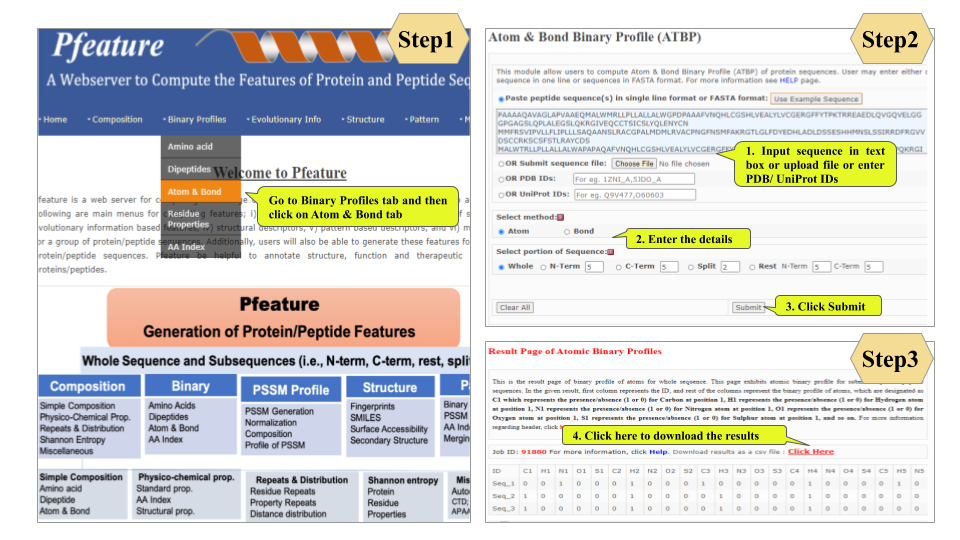
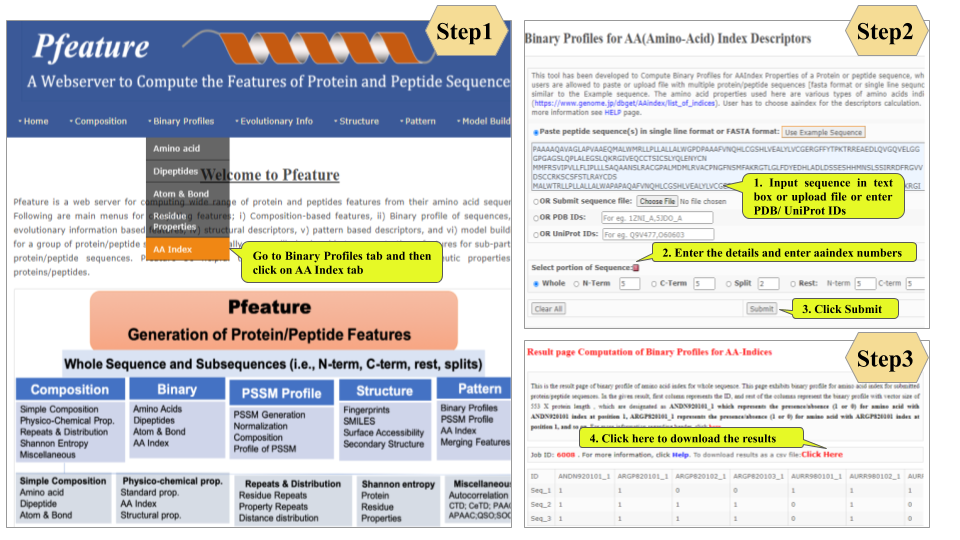
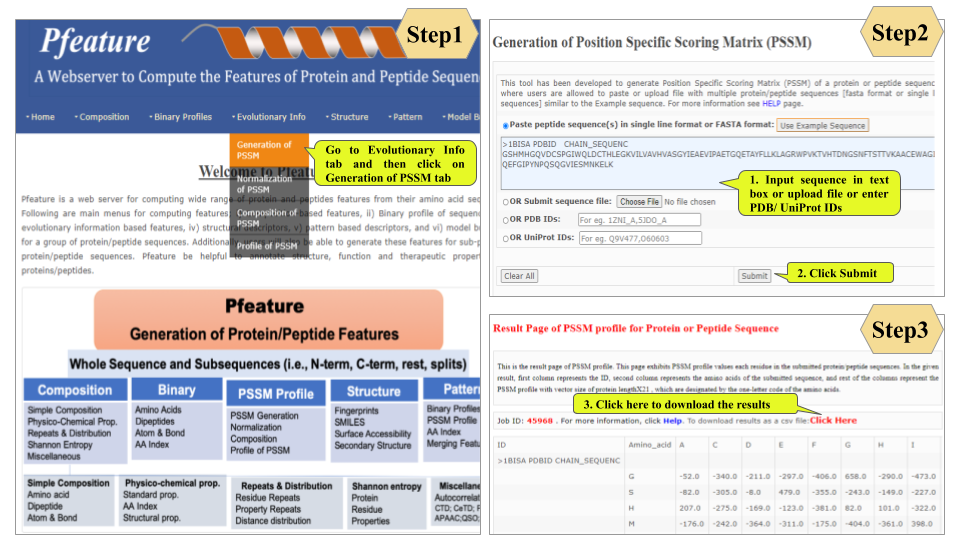
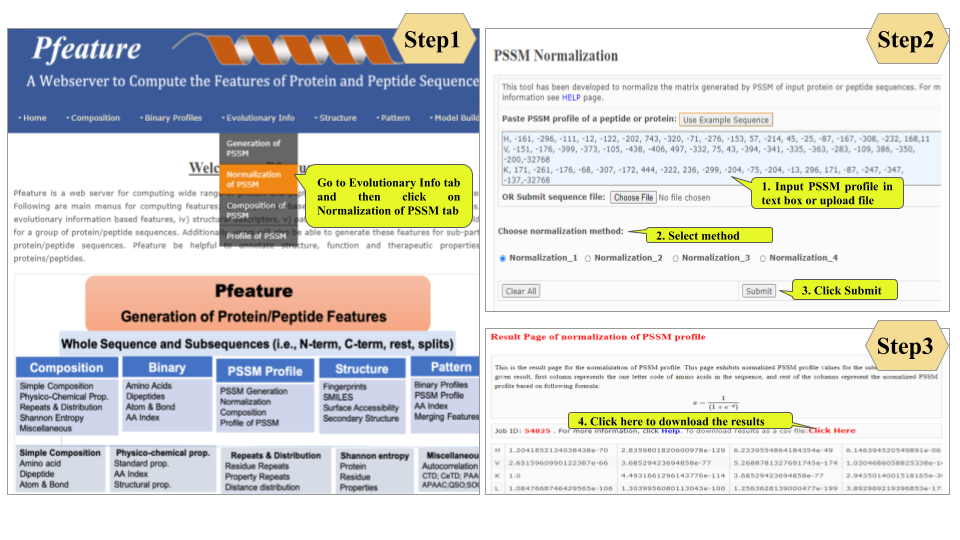
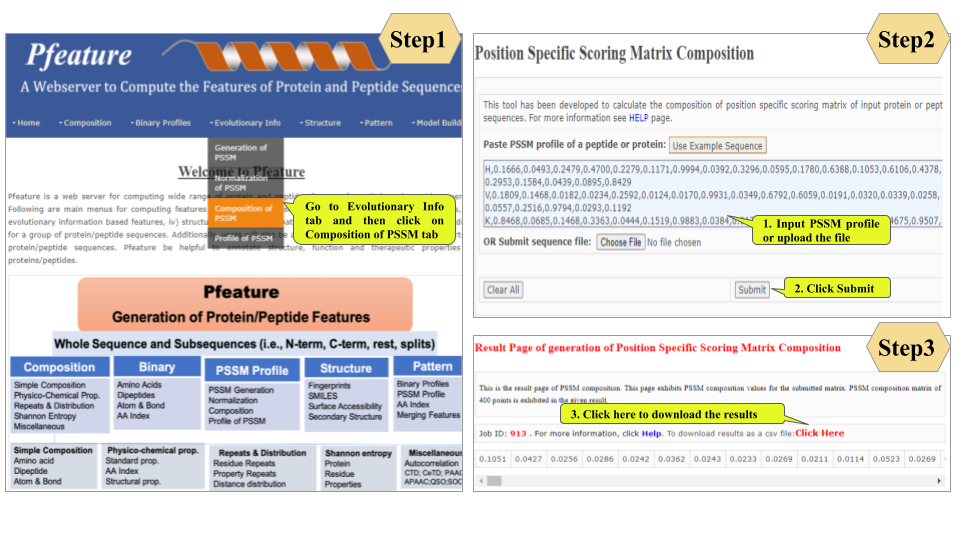
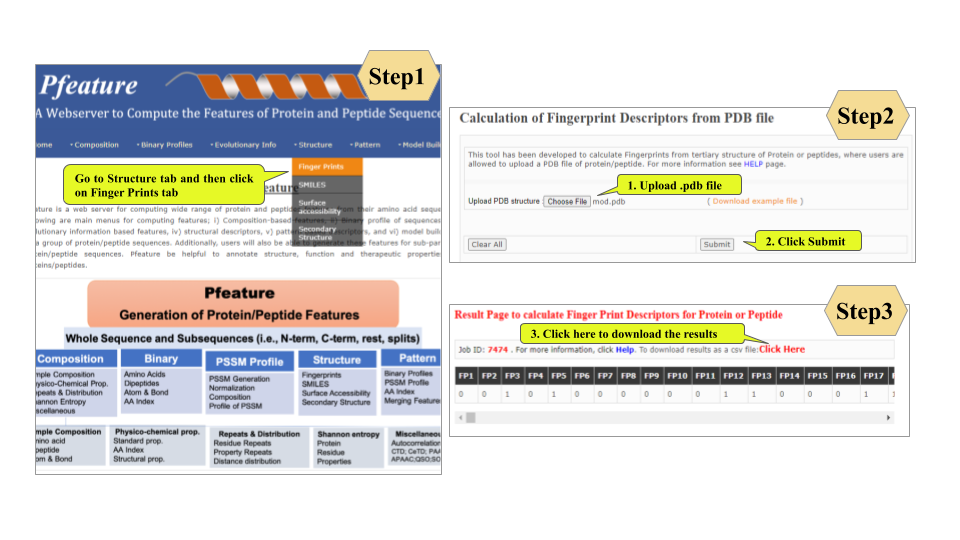
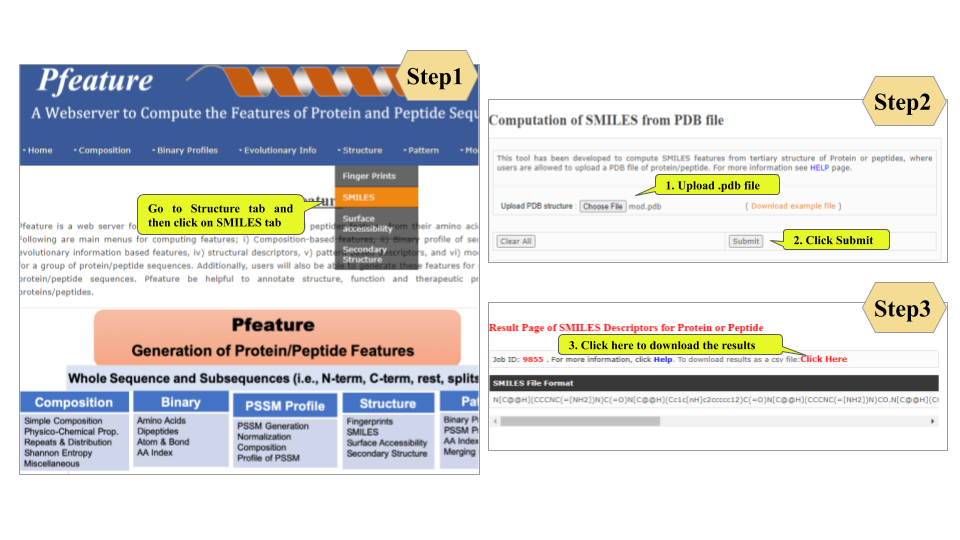
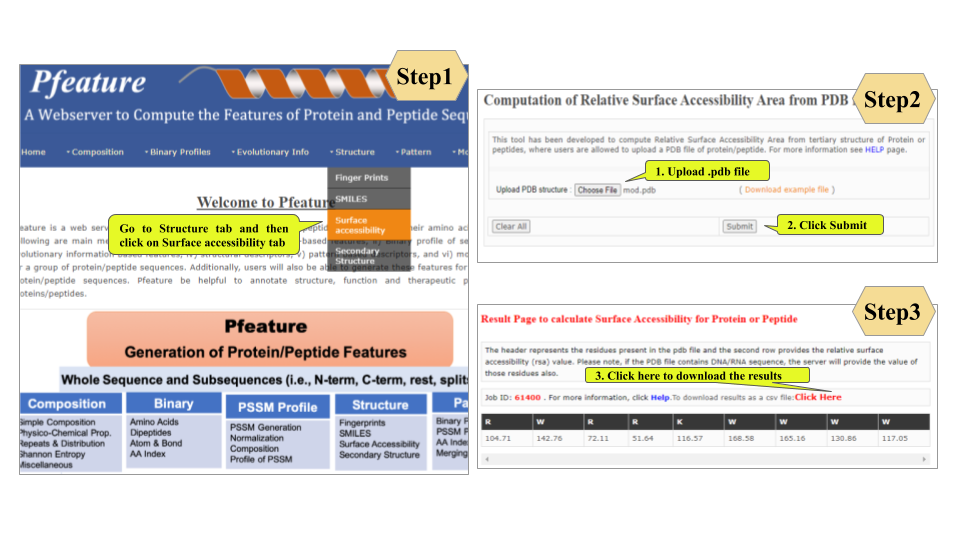
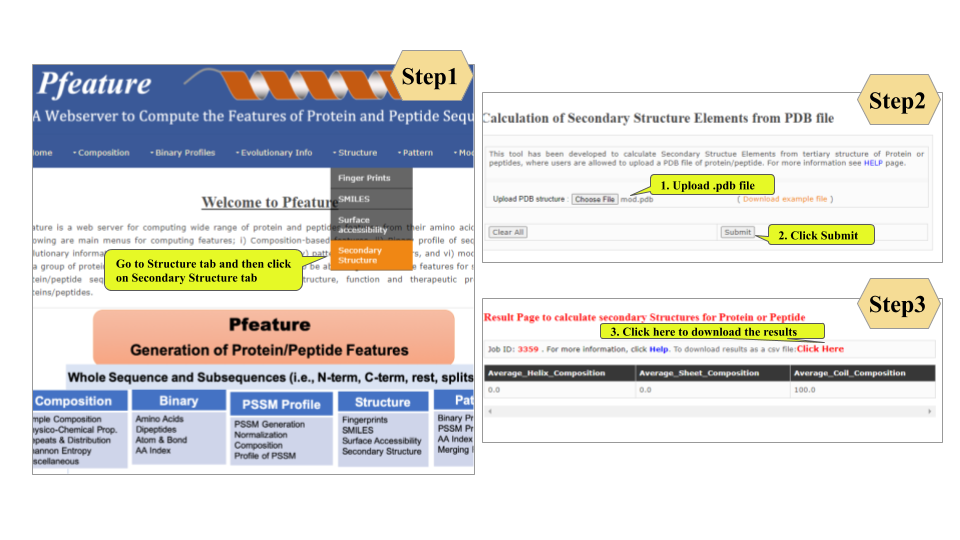
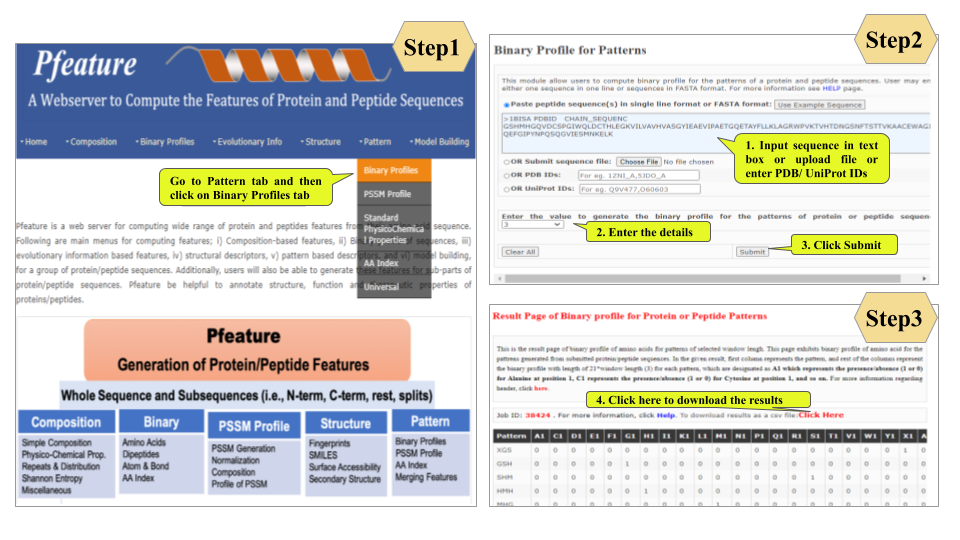
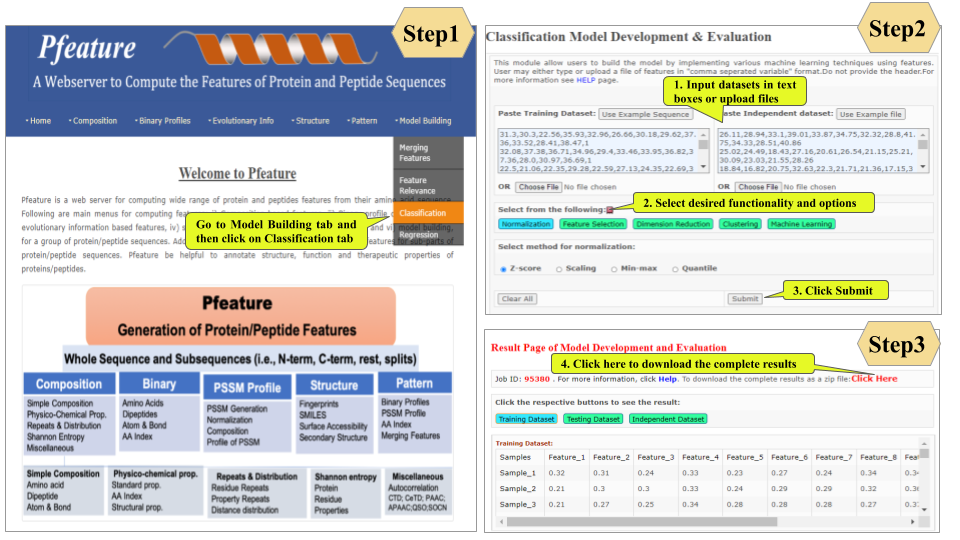
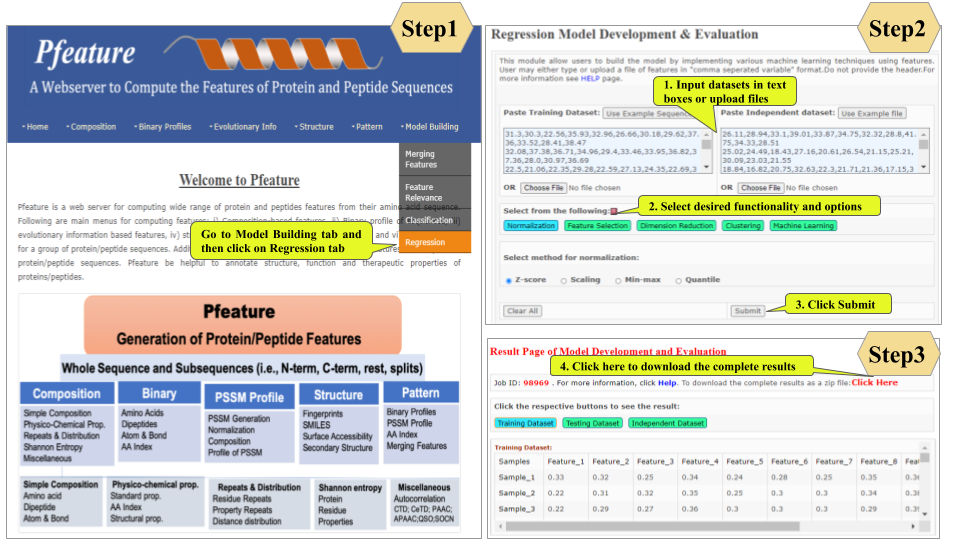
This page provides help to the users of Pfeature to compute the features of protein and peptide sequence. For each module which is implemented in Pfeature, here we provide the figures and description on how to use that module. Six general modules are provided with the respective.
To download the complete manual click  |
Frequently Asked Questions (FAQs) |
|
Q1. What is PFeature? Ans. Pfeature is a comprehensive webserver, which integrate numerous modules to compute most of the protein features that have been discovered over the past decades. Moreover, it allows to train, test and evaluate the model developed using the generated features. In this Webserver, we have integrated modules to generate six major categories of protein/peptide features: i) Composition-based features, ii) Binary profile of sequences, iii) Evolutionary information based features, iv) Structural descriptors, and v) Pattern based features vi) Model Building; for a group of protein/peptide sequences. Q2. Why PFeature? Ans. It is a complete package which includes steps from the generation of features to model building. |