SEARCH |
SIMPLE SEARCH
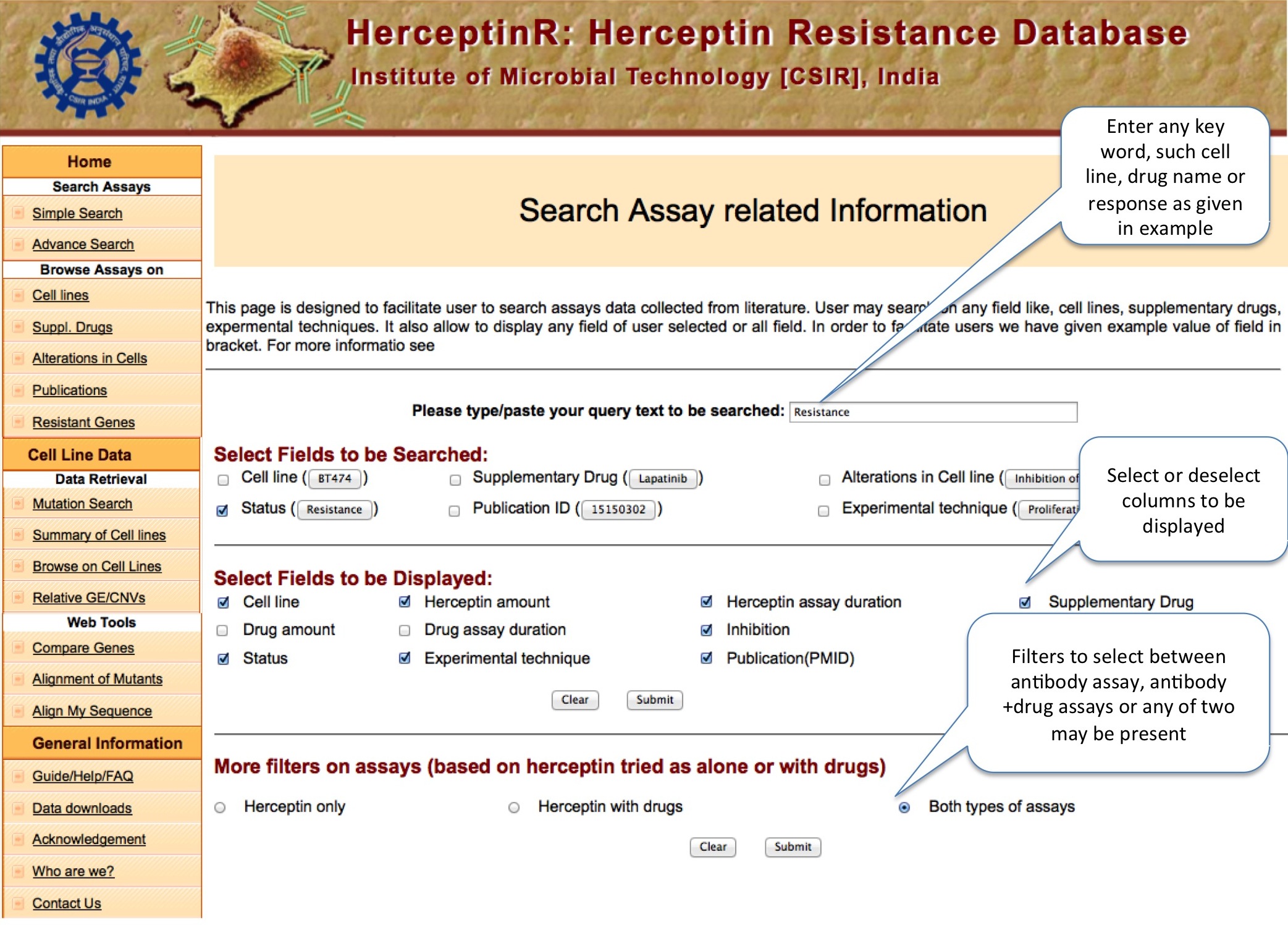
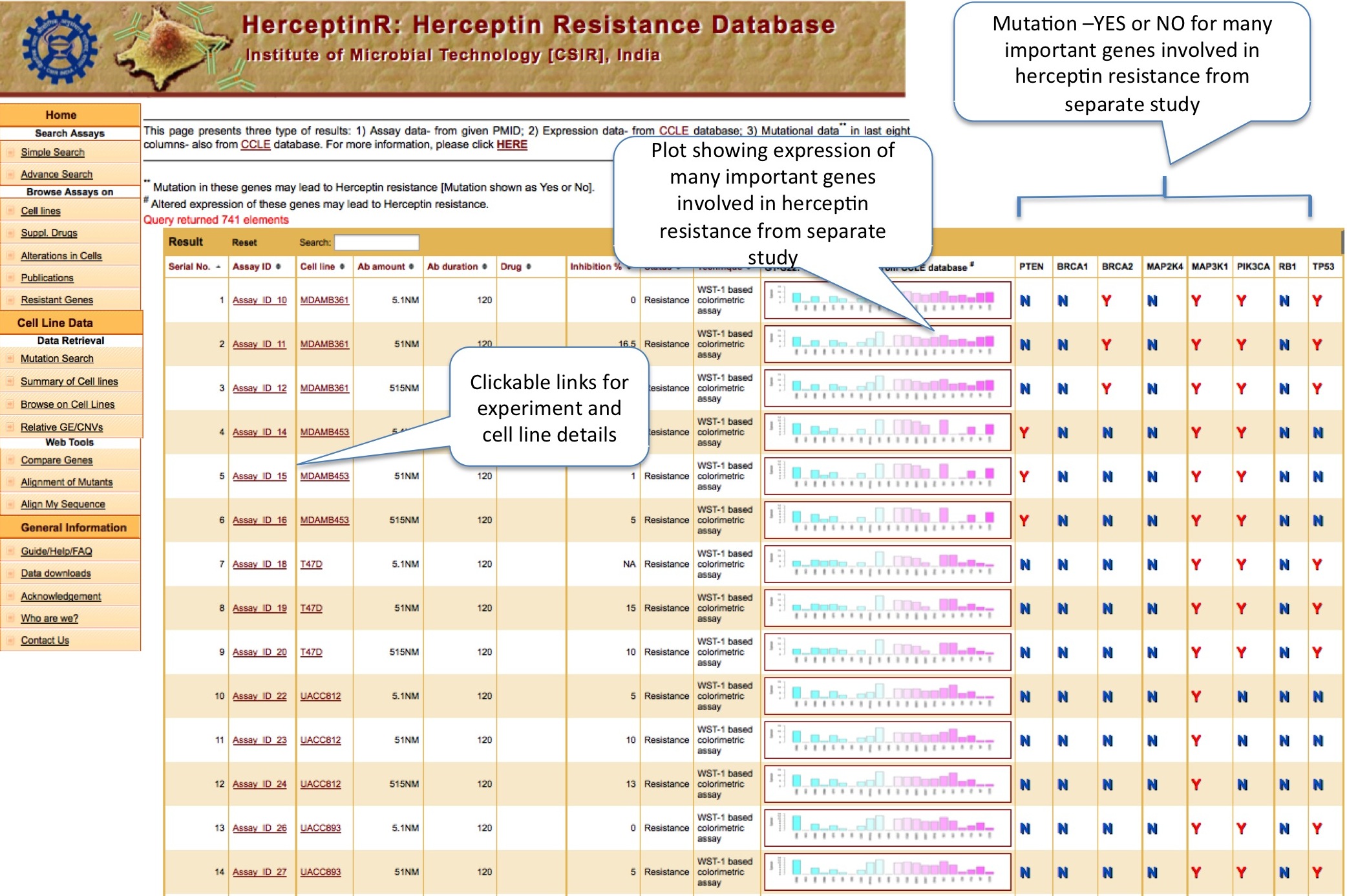
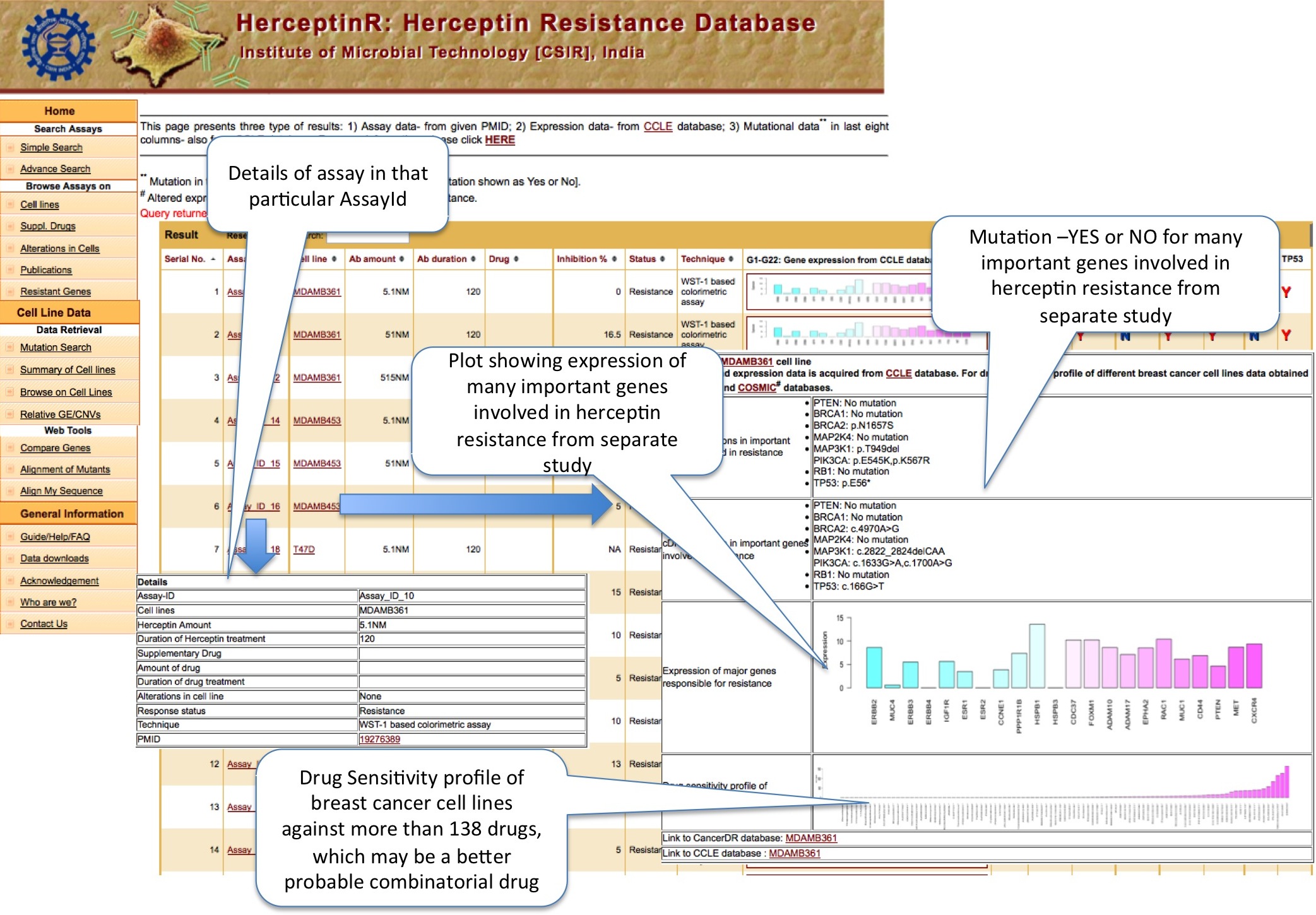
Simple search option enables user to search different assays performed with Herceptin with and without supplementary drug. By selecting particular cell line or status like resistance or sensitive, user can explore all those assays. To lessen the number of display column, user can select the desired column. If user want assays done with Herceptin only or only those which involves both Herceptin and drug, he/she can select 'more filters on assay' option. The display present the results in two different sections. First half of result shows the assay details and second half of result shows the genomic (expressional and mutational) status of some important genes. The plot shows expression of panel of 22 genes which are reported in literatures to be involved in Herceptin resistance. Next to the plot, column of eight genes in the form of YES and NO table, which shows whether these genes are mutated (Y) or not (N) in particular cell line. These genes are also hypothesized in litrature to be contributing in Herceptin resistance. Genomic data is acquired from separate study of CCLE database.
|
|
|
|
|
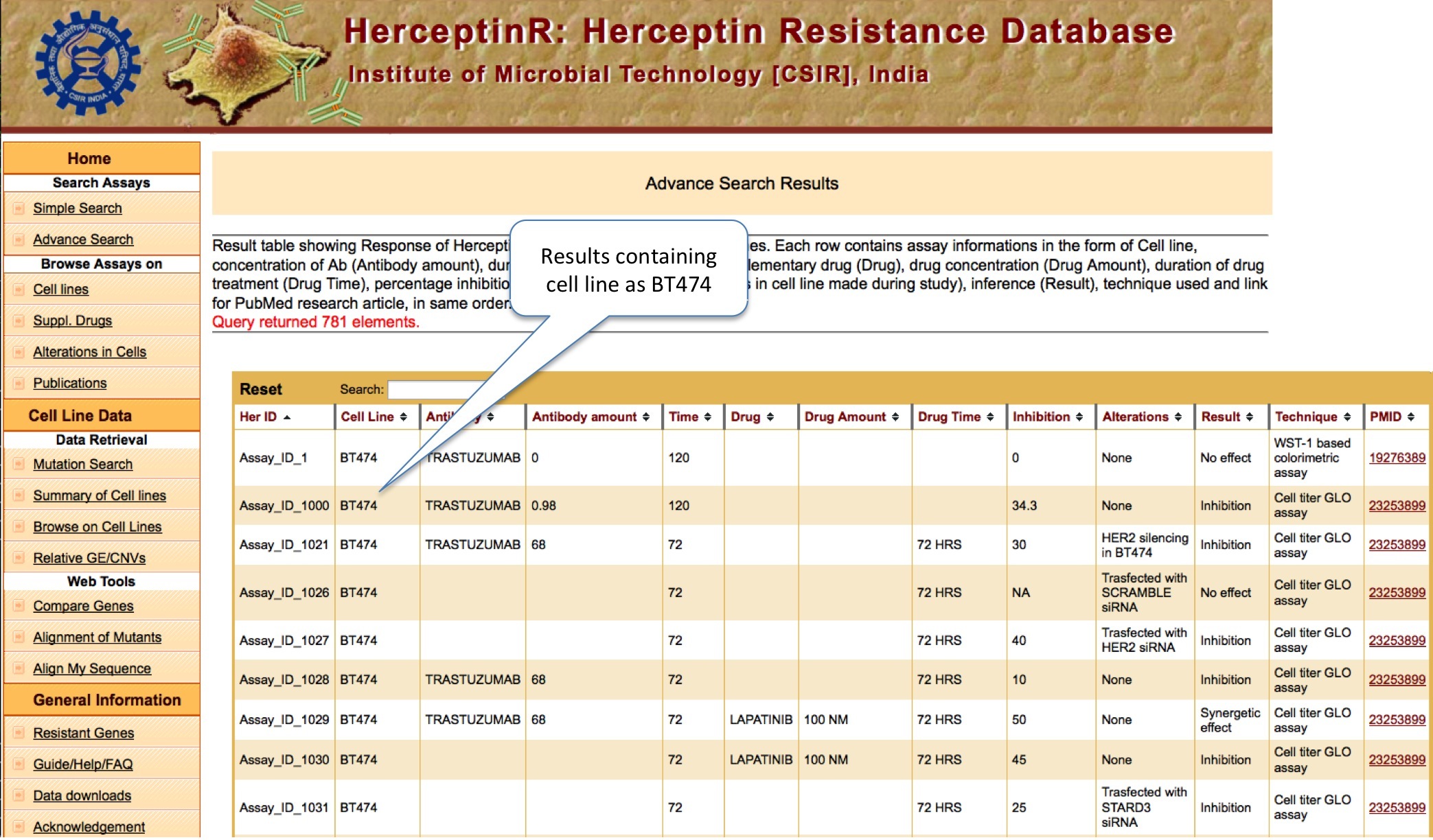
ADVANCE SEARCH
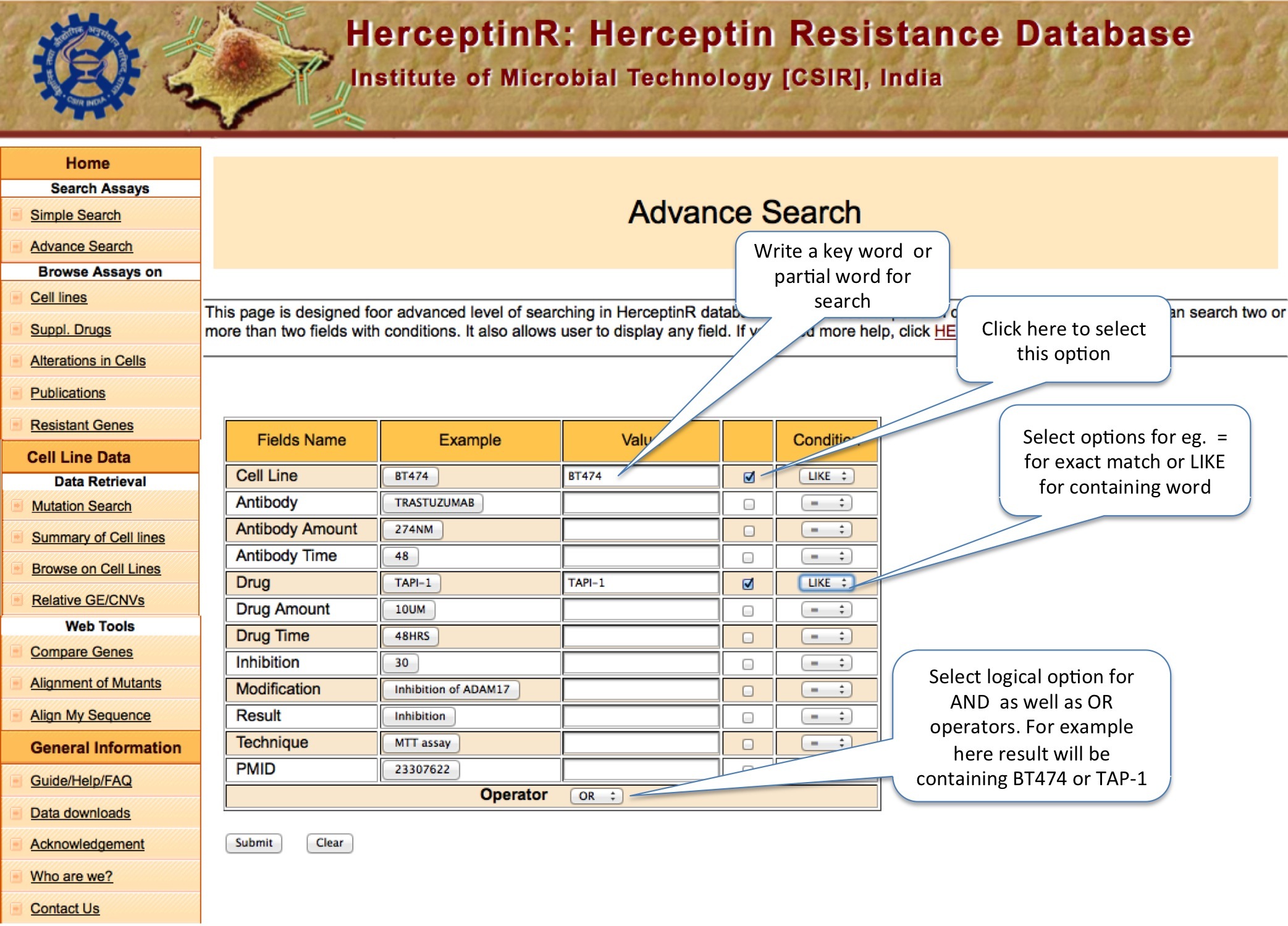
This option is meant for extensive search with logical operators like AND and OR and exact or containing (like) matching.
|
|
|
|
BROWSE |
|
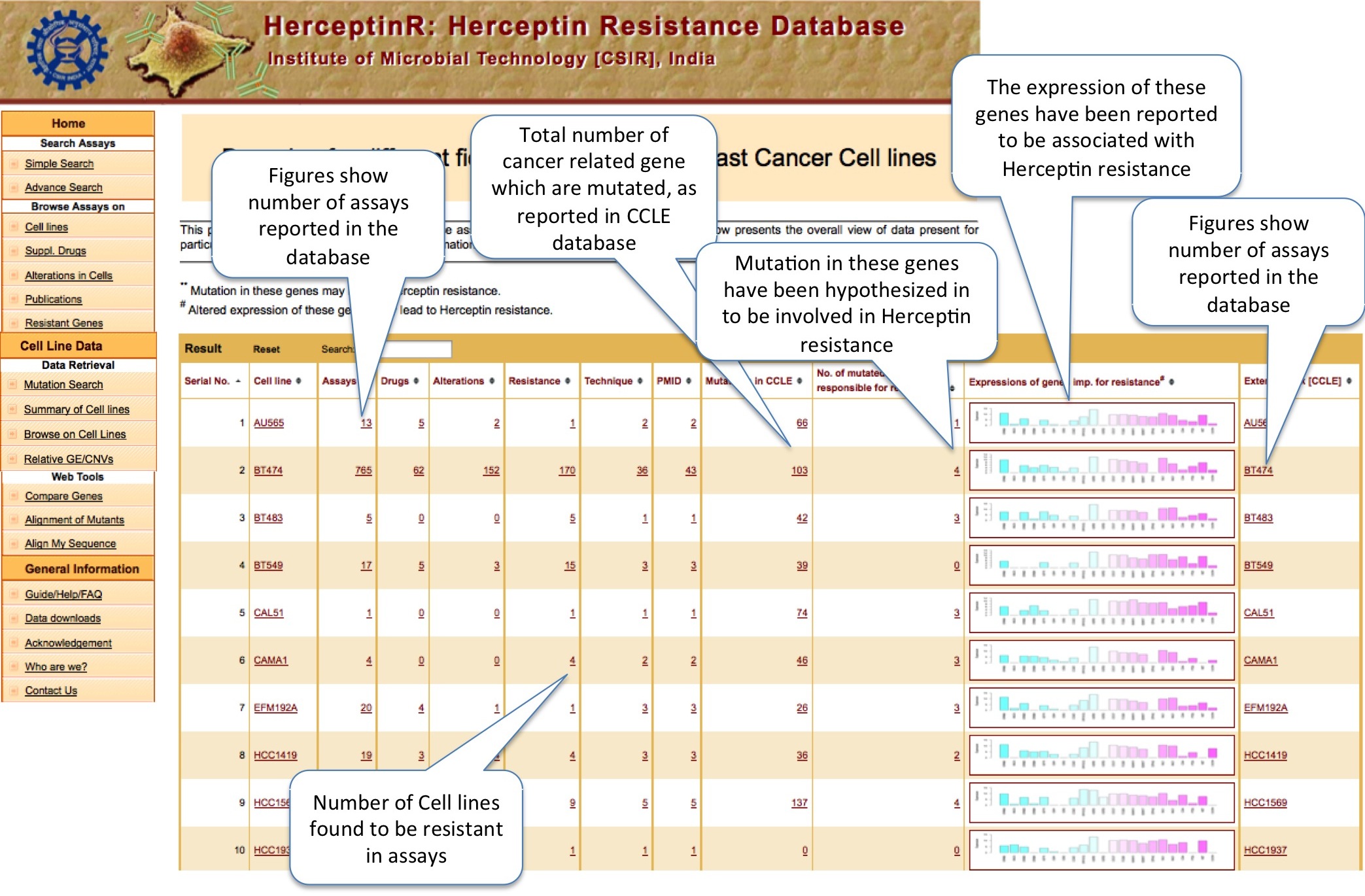
CELL LINES
Here user can find out the important information about cell lines e.g. number of assays, alteration, their mutation status, expression status, important links etc.
|
|
|
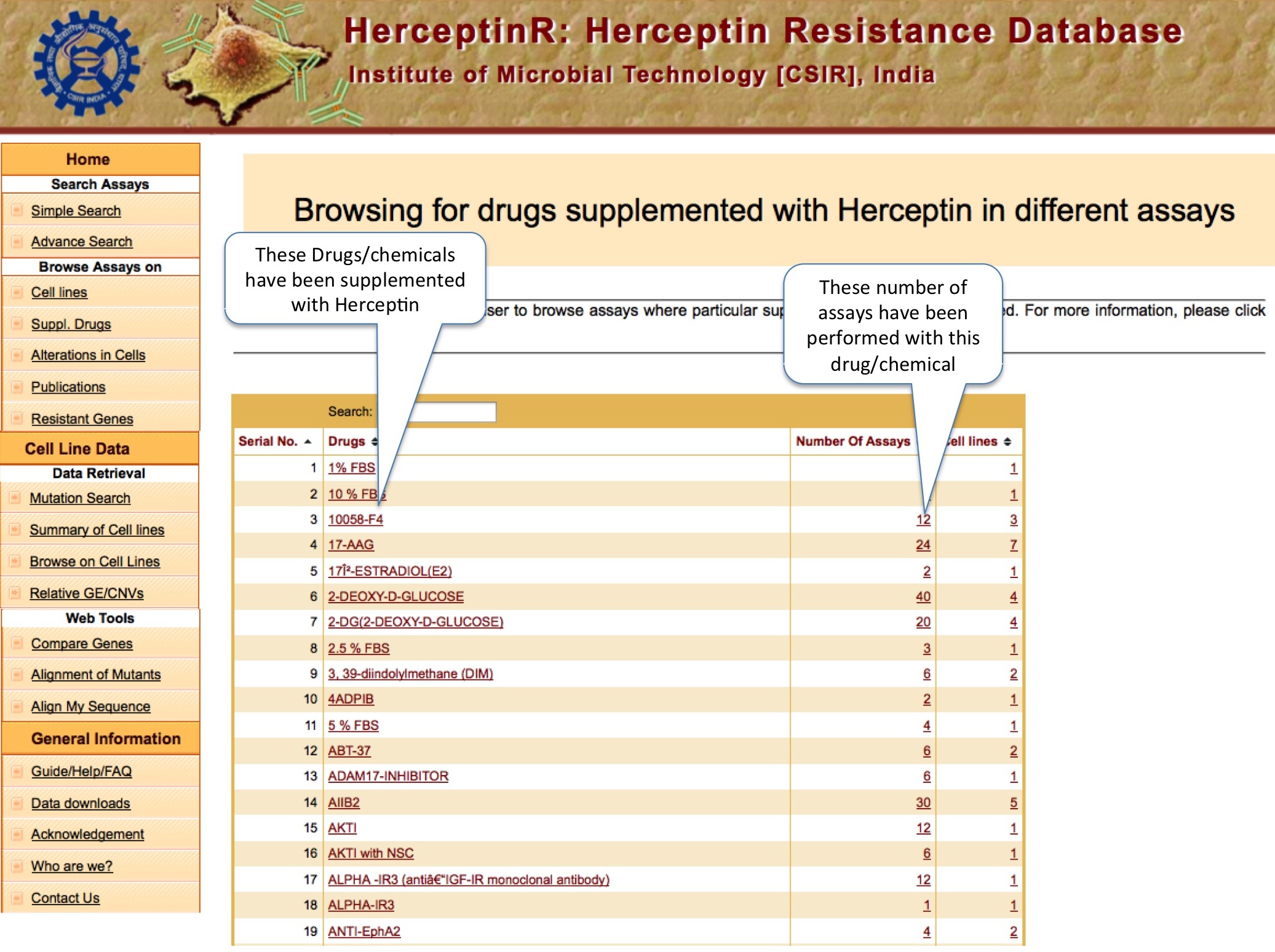
DRUGS
This tool will help users to find out the supplementary drugs used in various assays available in HerceptinR and number of cell lines.
|
|
|
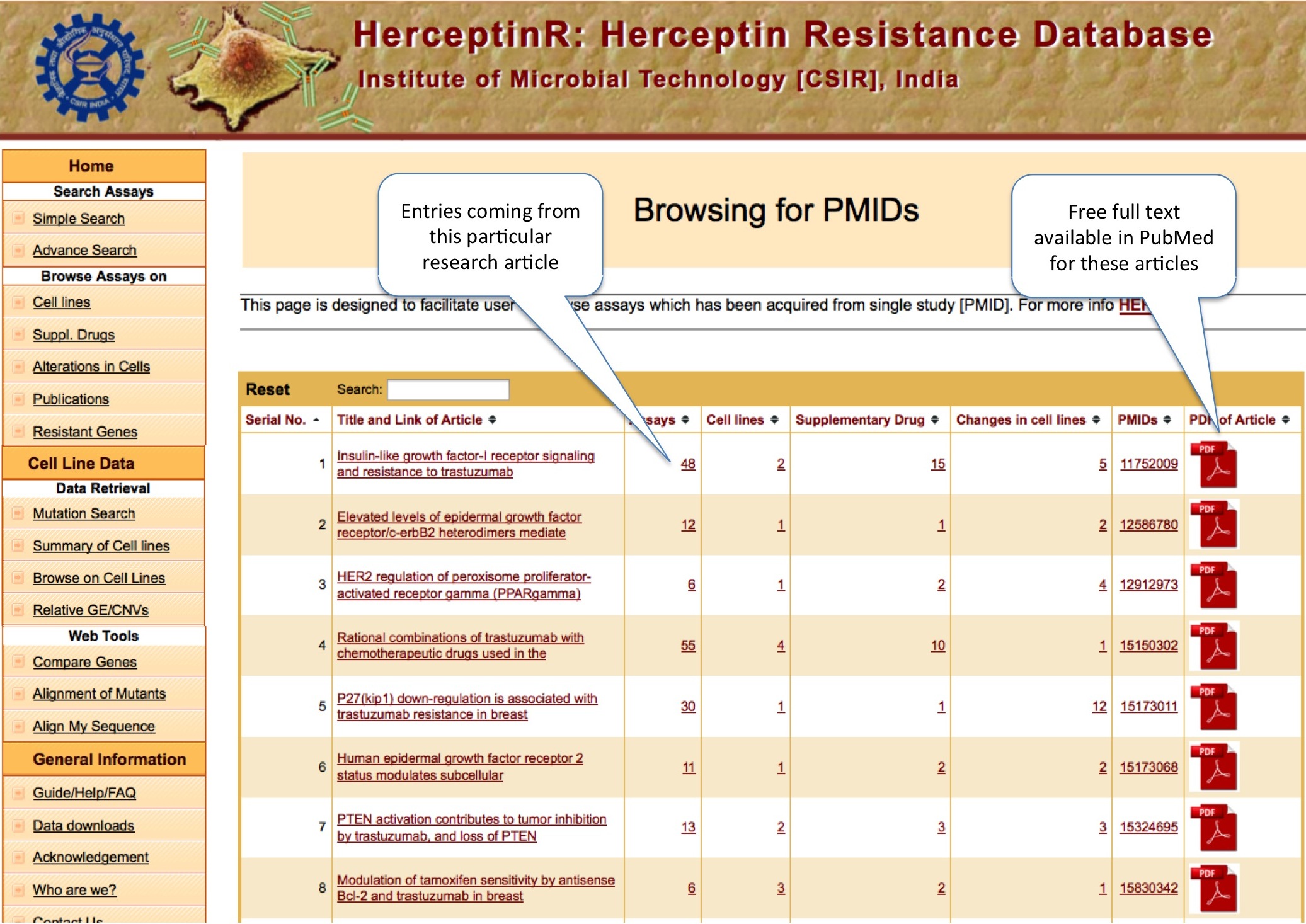
PMIDs
Here user will find out the literature information of different assays in terms of title of the paper. User can also download the pdf format of the research articles.
|
|
|
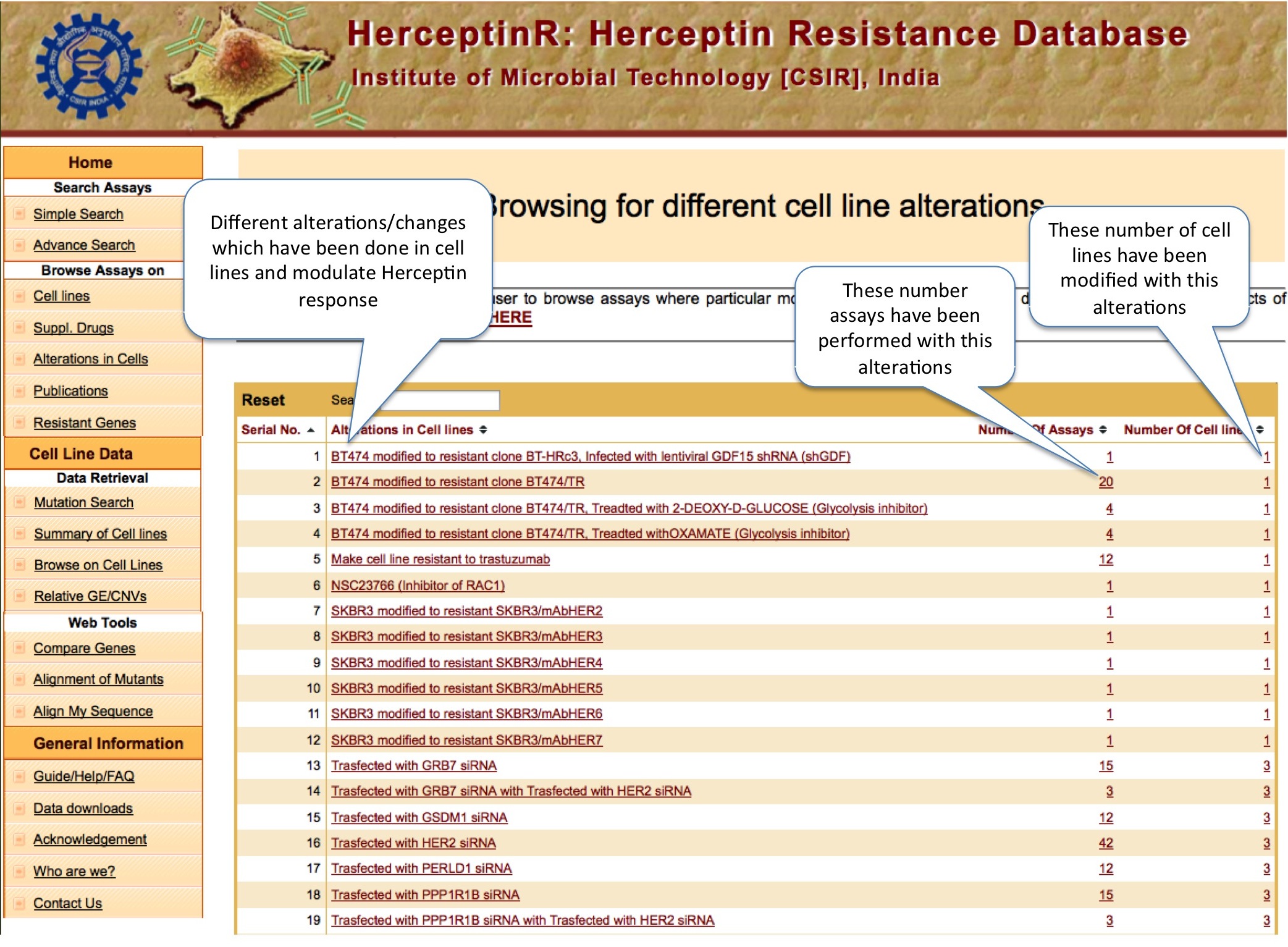
ALTERATIONS IN CELL LINES
There are ~337 types of alterations, which have been done in BCCs of HerceptinR database. Each row provides the alteration, number of assays performed with that alteration and number of cell lines with this alteration, in our database.
|
|
SEQUENCING DATA |
|
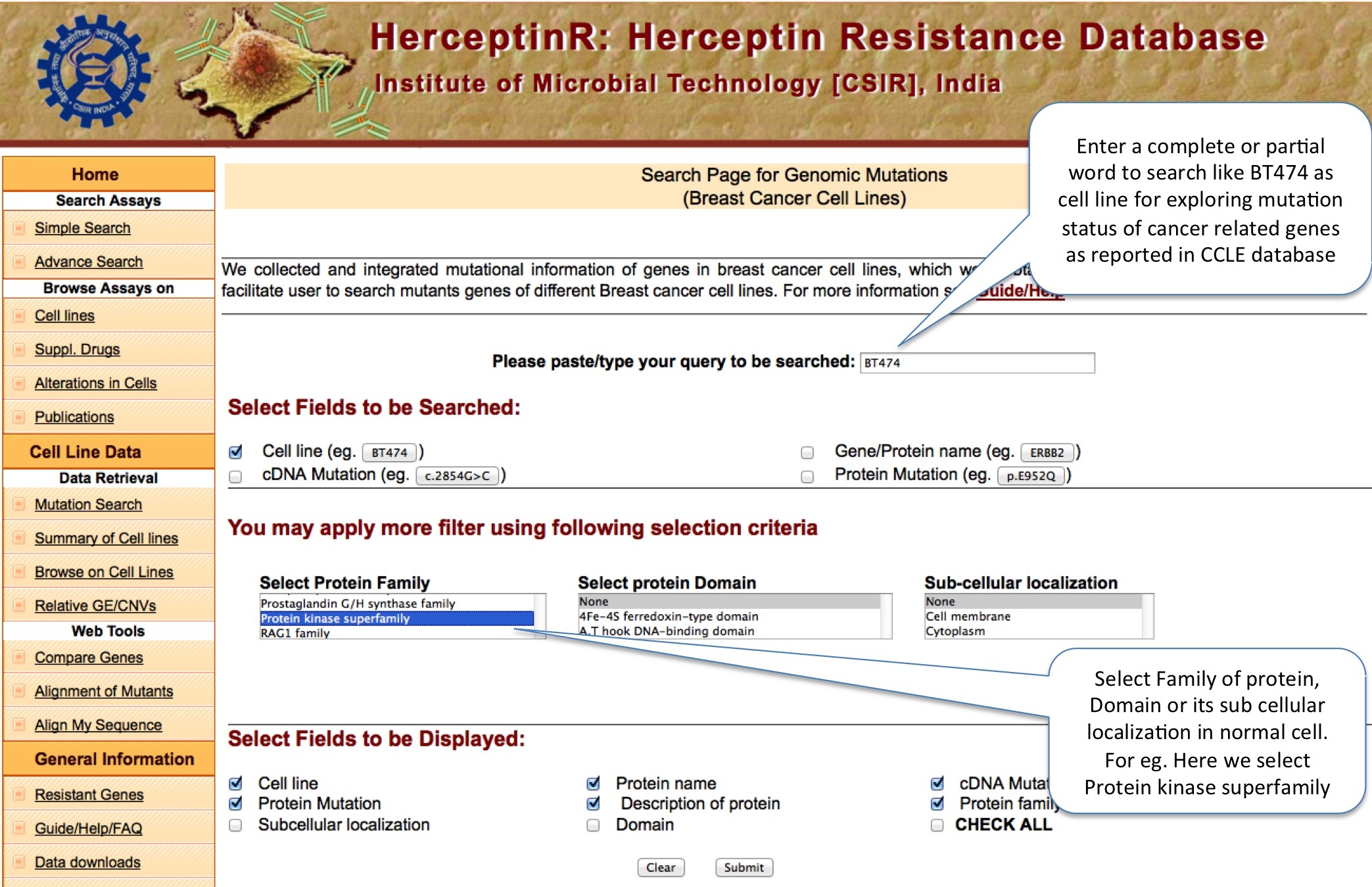
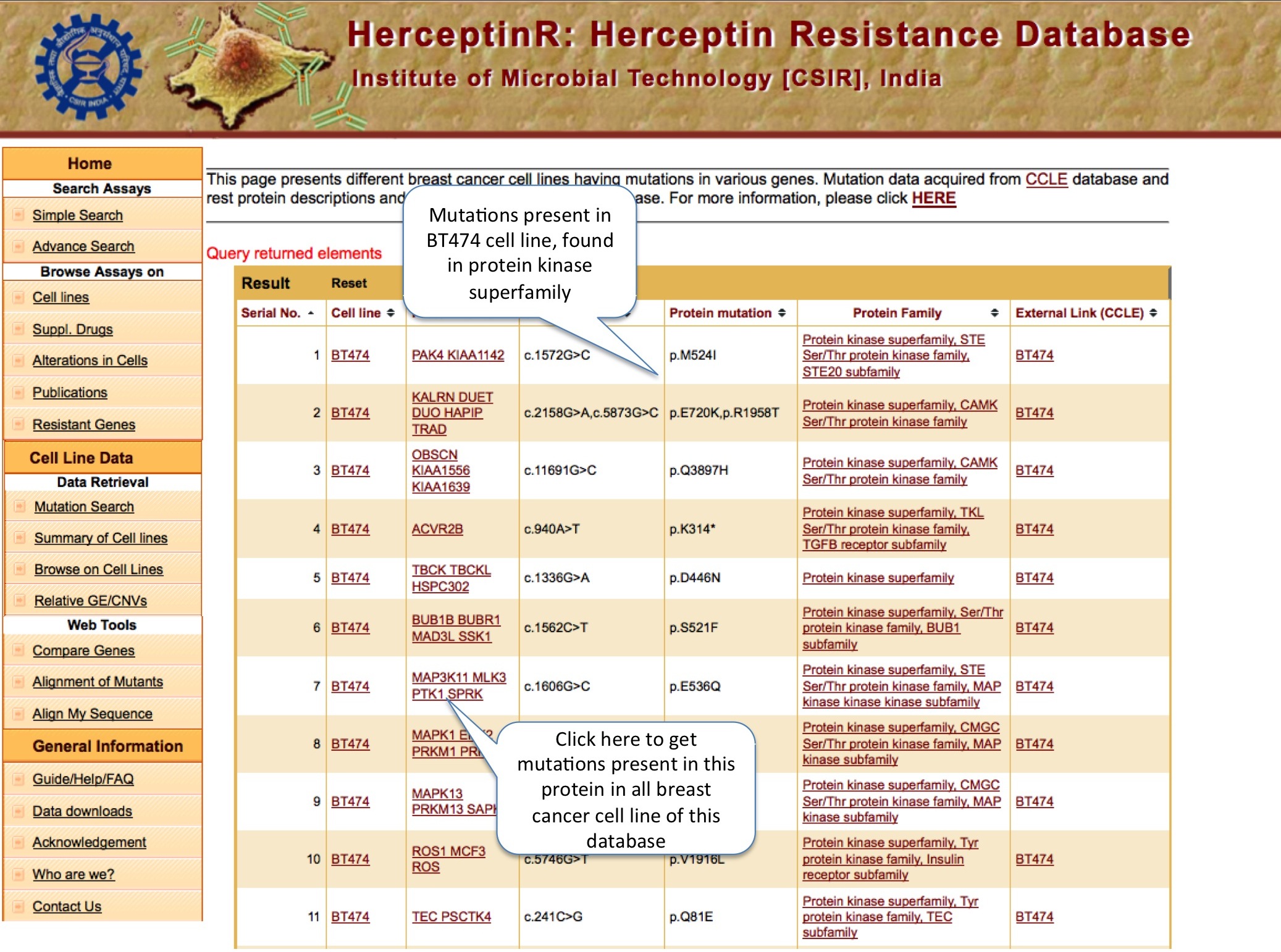
MUTATION SEARCH
Here user can search the type of mutation present in cell lines both at the cDNA and protein level. Along with this, user can find out the information about protein family, domain and subcellular localization.
|
|
|
|
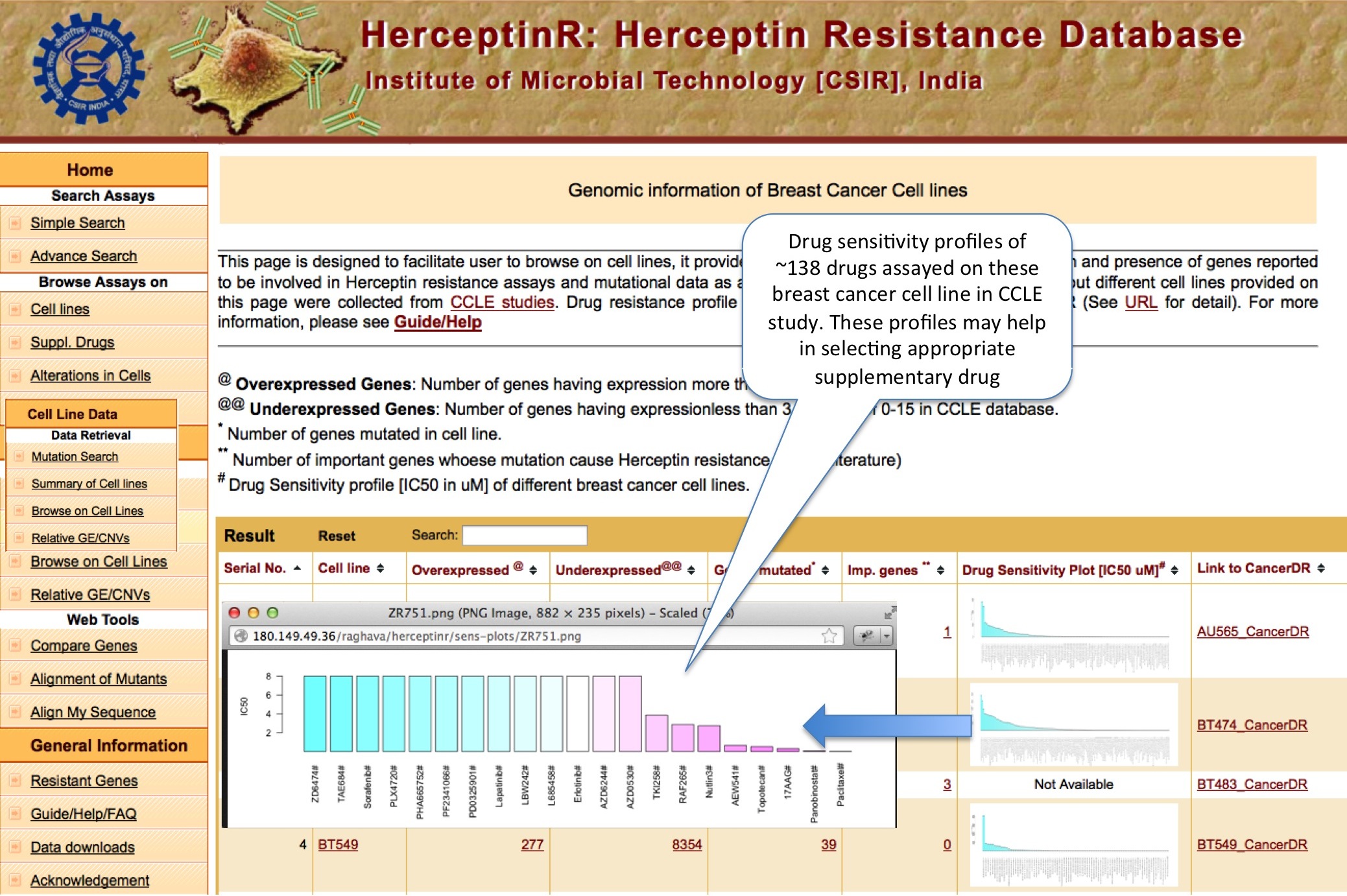
SUMMARY OF CELL LINES
Here various important information are assembled for all the cell lines in terms of gene expression, mutation, mutation responsible for resistance and drug sensitivity profile of ~138 drugs.
|

|
|
|
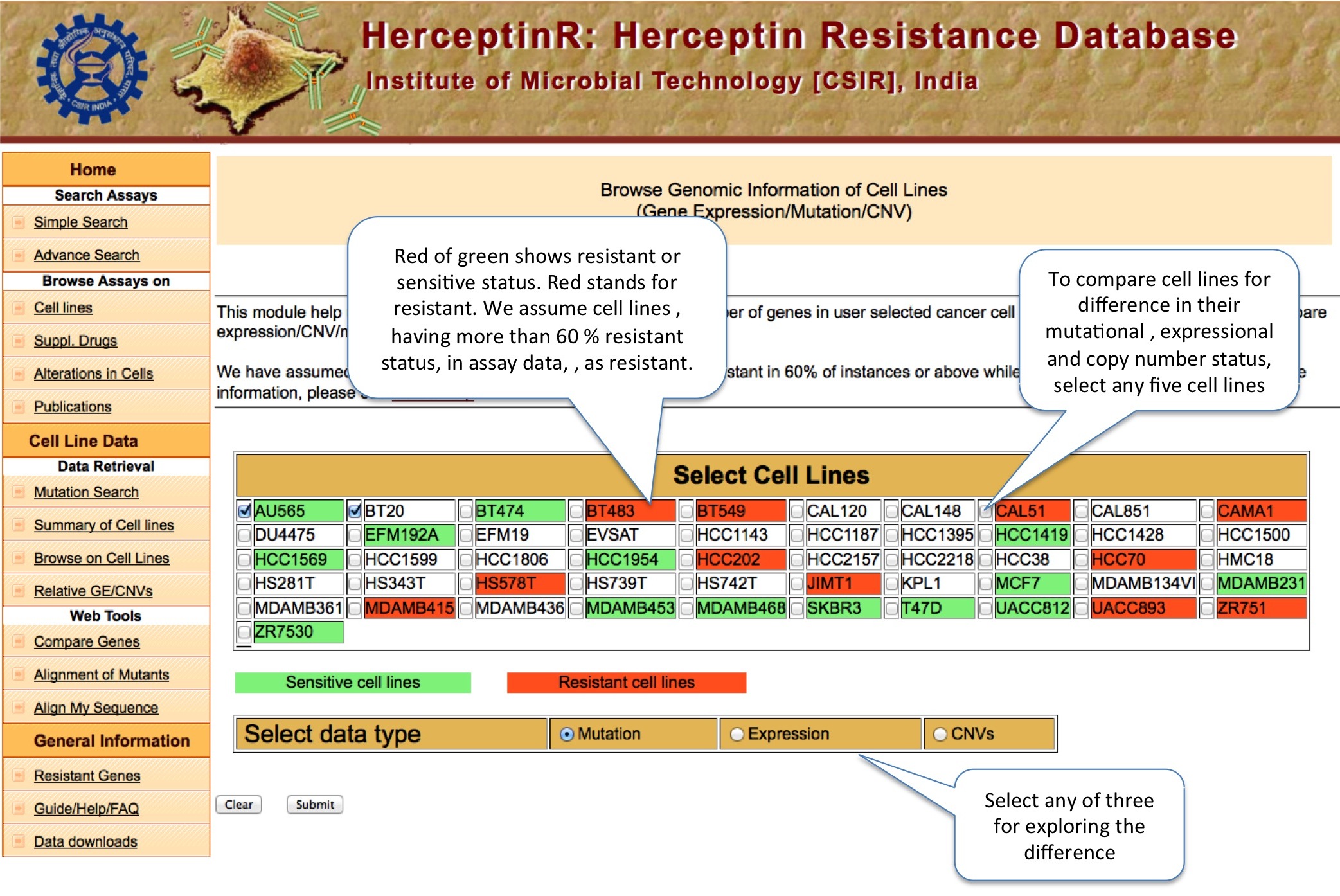
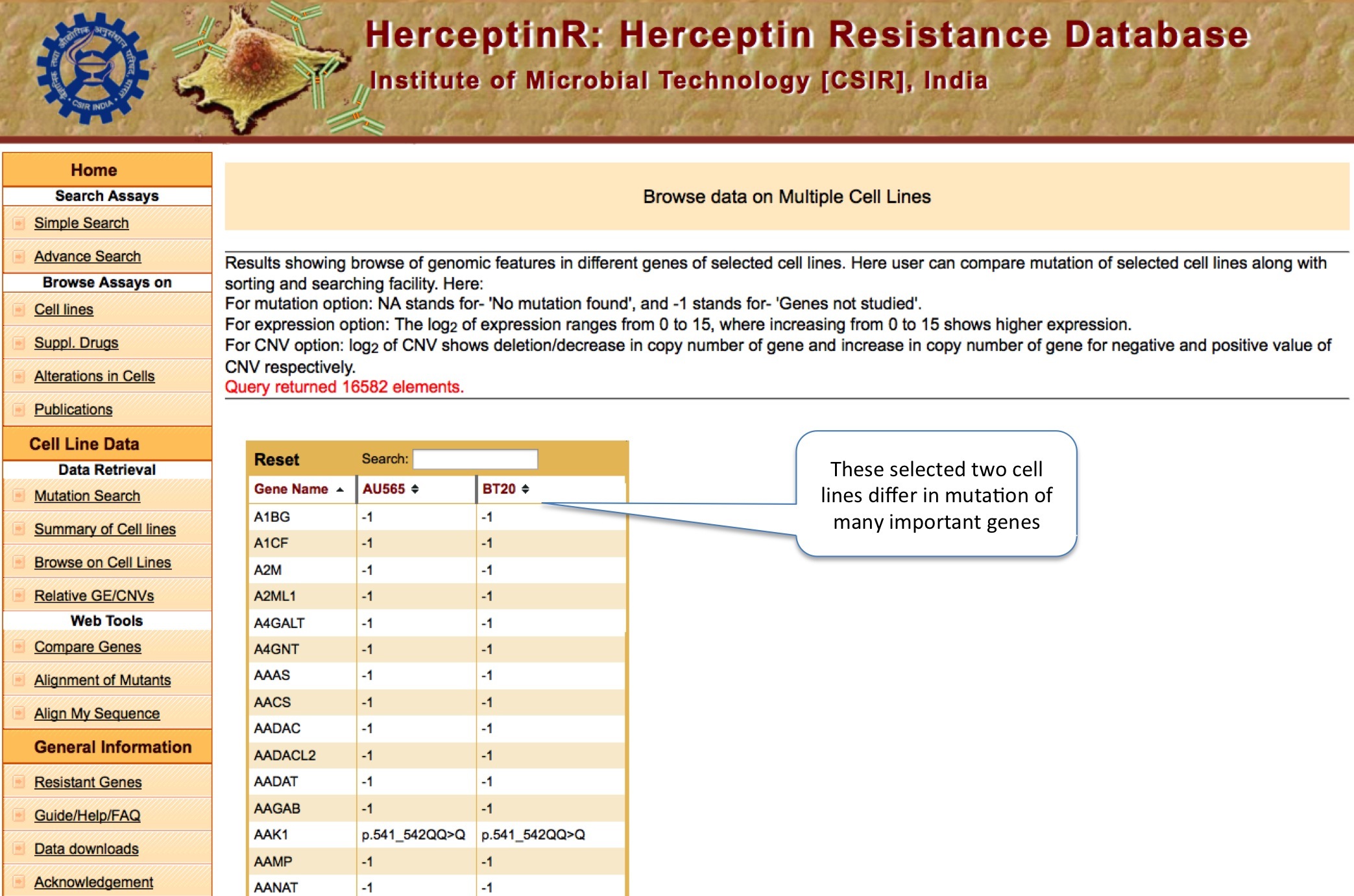
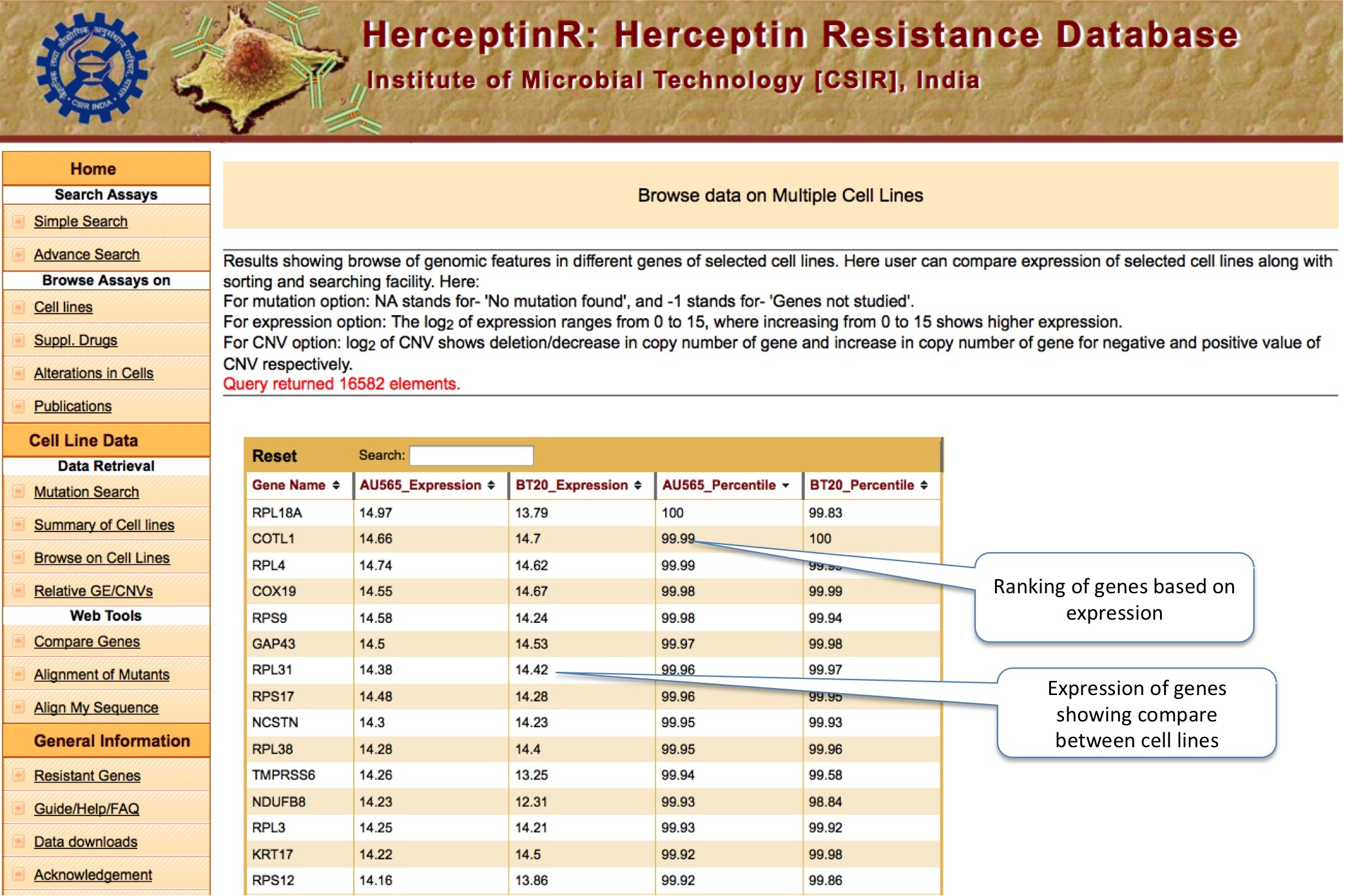
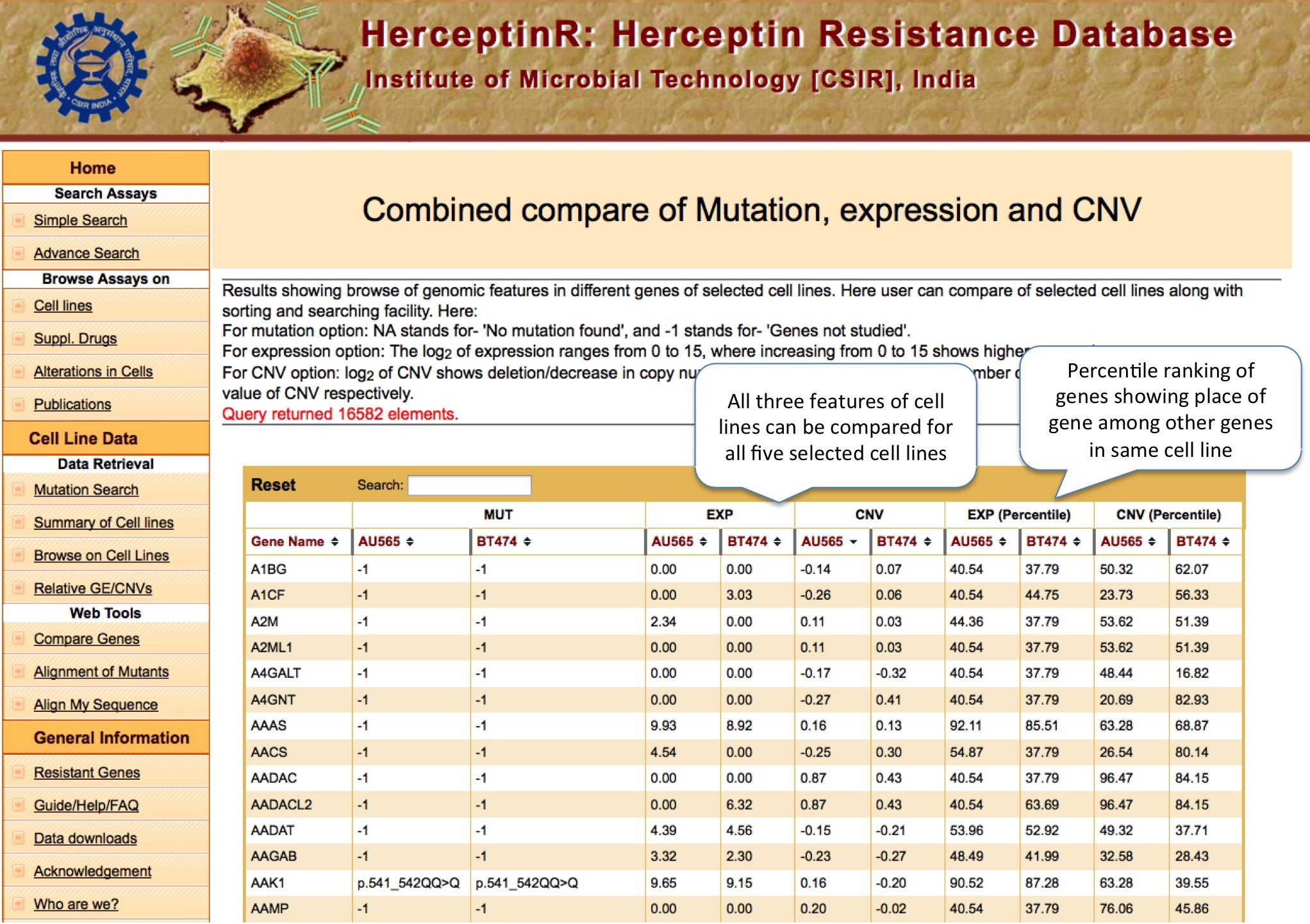
BROWSE ON CELL LINE
This tool allow user to compare the mutation, copy number variation and gene expression status of thousand of important genes for up to five cancer cell lines at the same time. The Percentile Rank of a gene shows the rank of gene among different genes of same cell line bsed on CNV or expression. The formula used is: Pg1 = (E+B)/N ; where P is percentile rank for gene g1 , E is number of genes having equal expression/CNV, B is number of genes having expression/CNV below g1 and N is total number of genes.
|
|
|
|
|
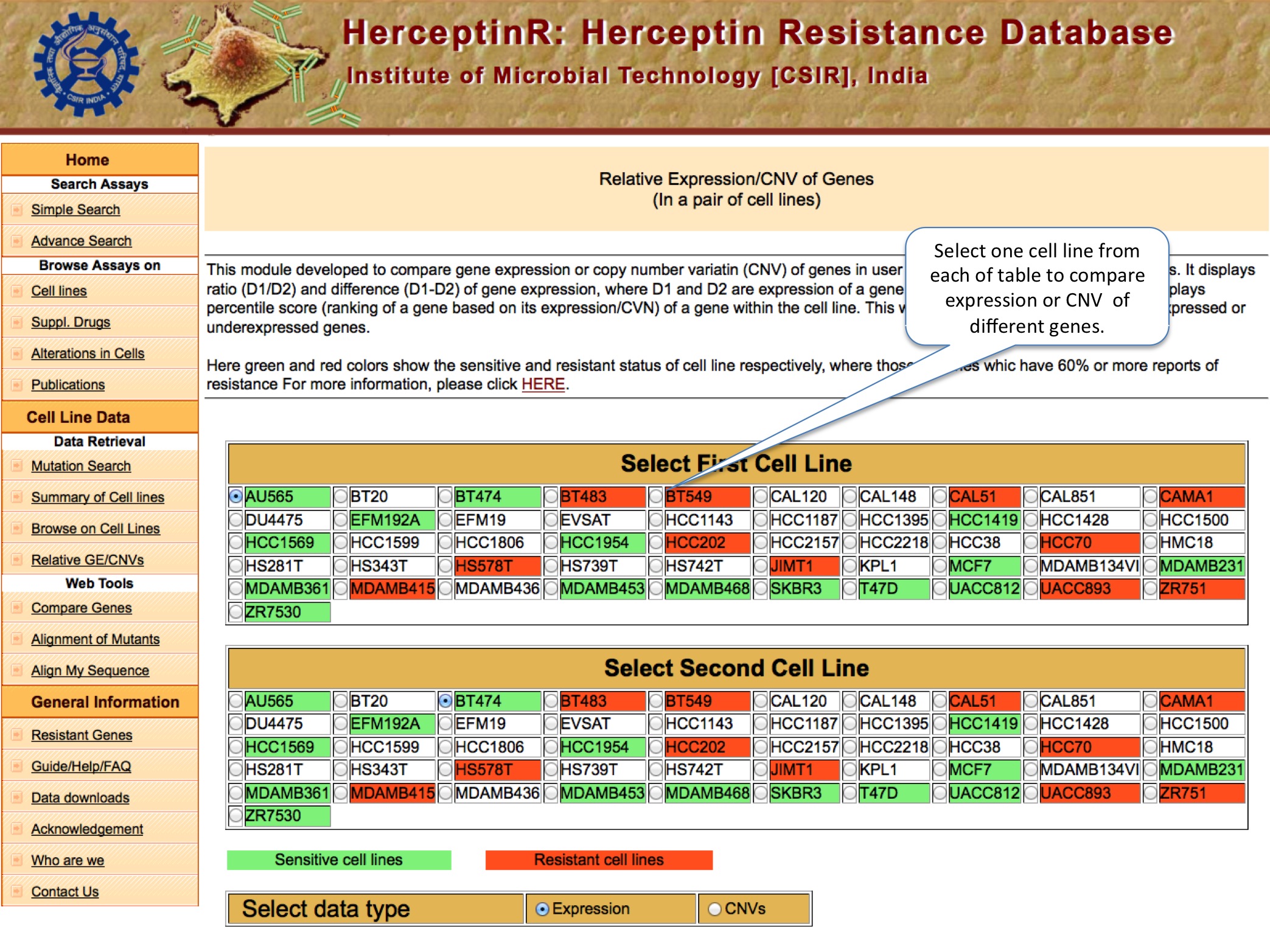
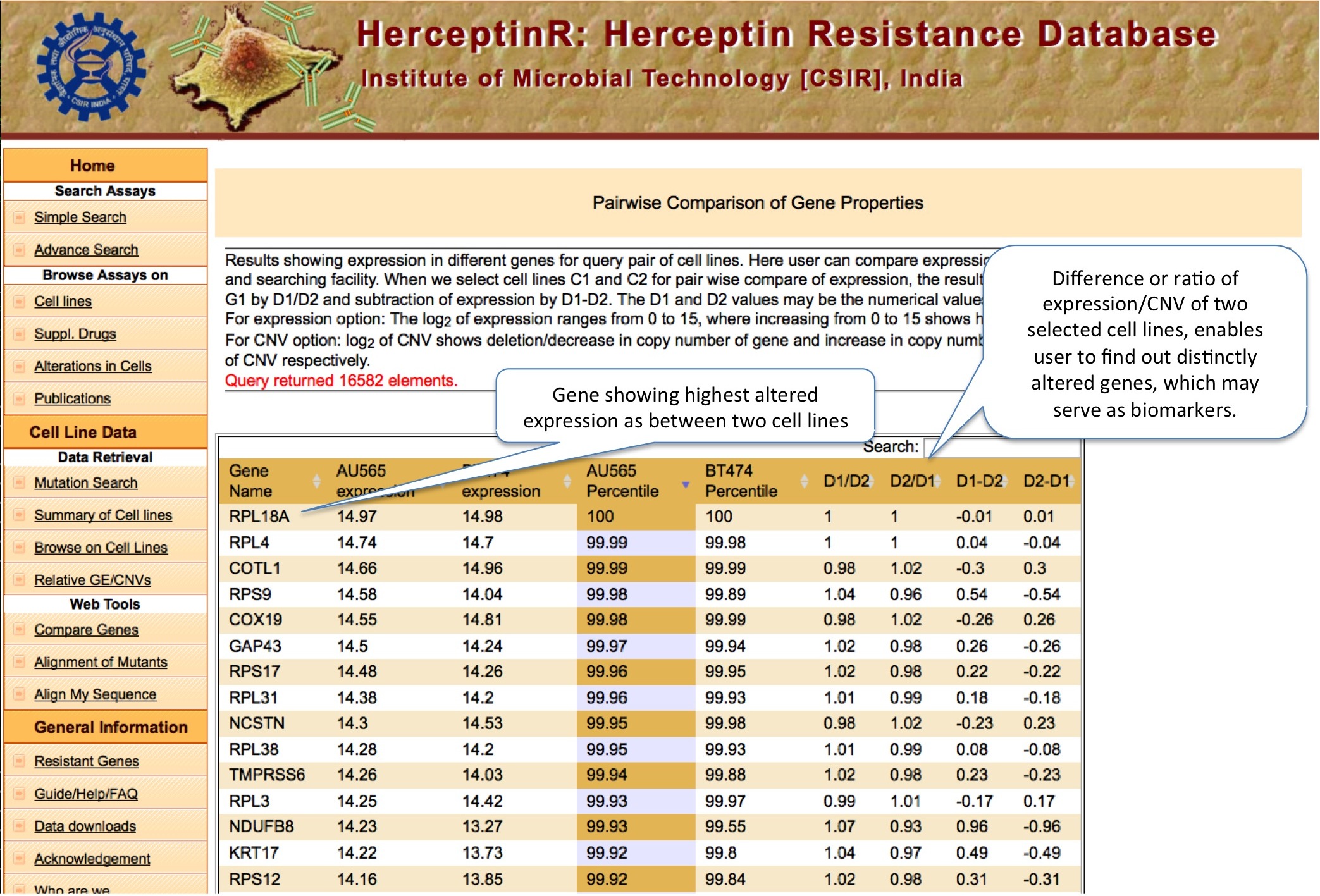
RELATIVE GE/CNV
Here user can do the comparative analysis of copy number variation and gene expression of thousands genes in any two cell lines. The Percentile Rank of a gene shows the rank of gene among different genes of same cell line bsed on CNV or expression. The formula used is: Pg1 = (E+B)/N ; where P is percentile rank for gene g1 , E is number of genes having equal expression/CNV, B is number of genes having expression/CNV below g1 and N is total number of genes.
|
|
|
|
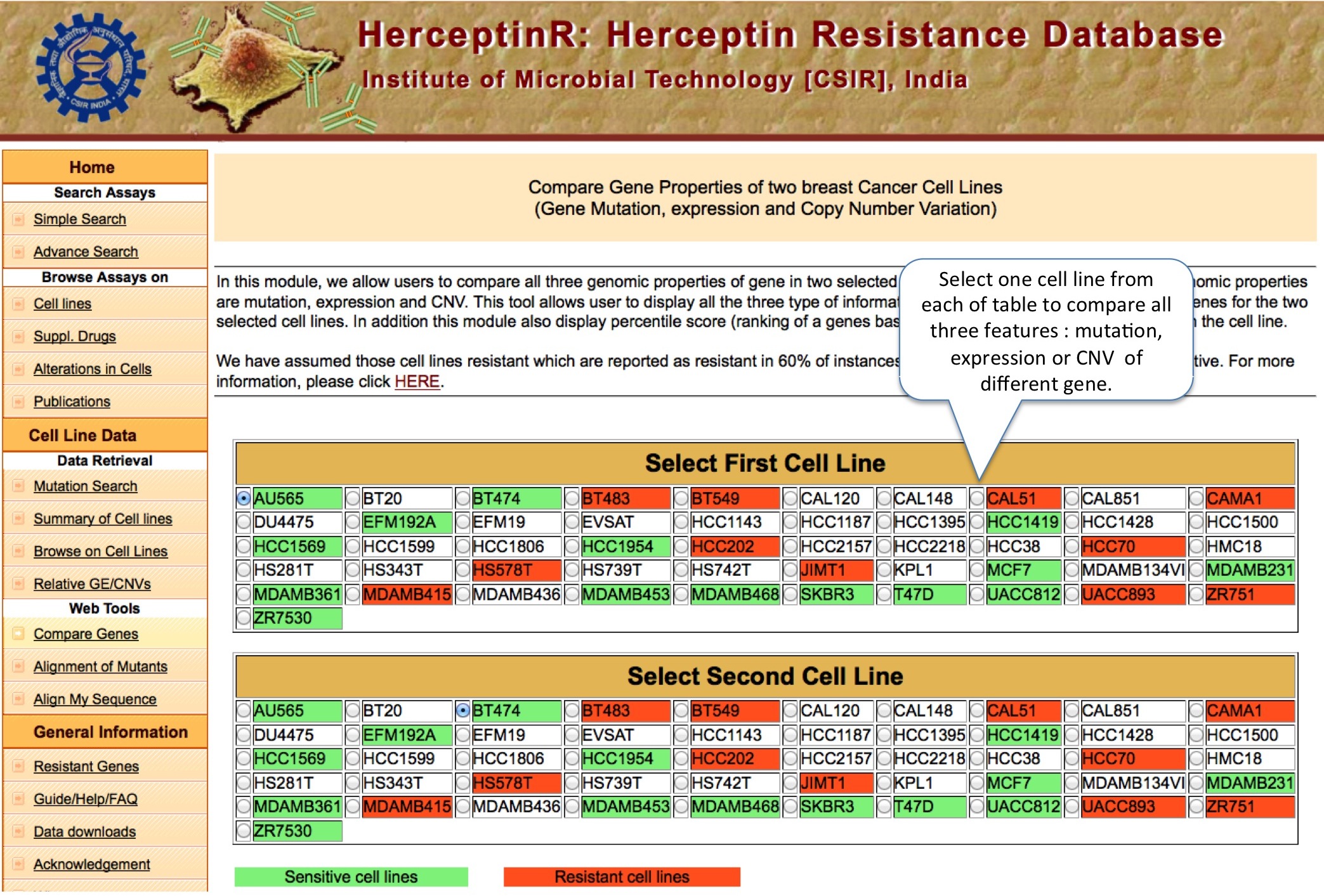
COMPARE GENES
Unlike above tools, here user can compare analyze the mutation, gene expression and CNV status of important genes at one place for any two cell lines. The Percentile Rank of a gene shows the rank of gene among different genes of same cell line bsed on CNV or expression. The formula used is: Pg1 = (E+B)/N ; where P is percentile rank for gene g1 , E is number of genes having equal expression/CNV, B is number of genes having expression/CNV below g1 and N is total number of genes.
|
|
|
|
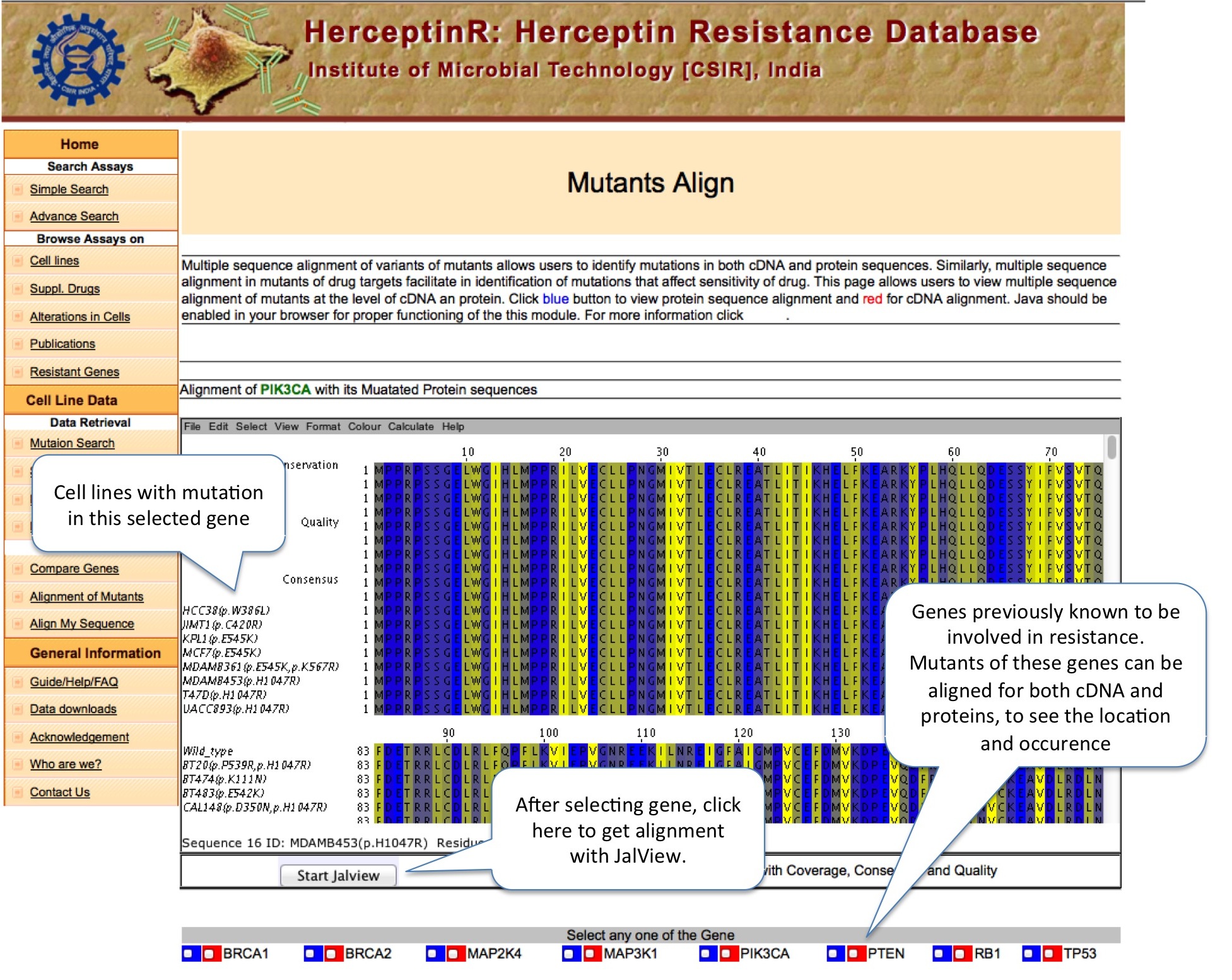
ALIGNMENT OF MUTANTS
This module allow users to view the sequence alignment of mutated gene or protein to find out the position of mutations. User can launch the Jalview by clicking on Start Jalview, which pop-up separate window to show alignment.
|
|
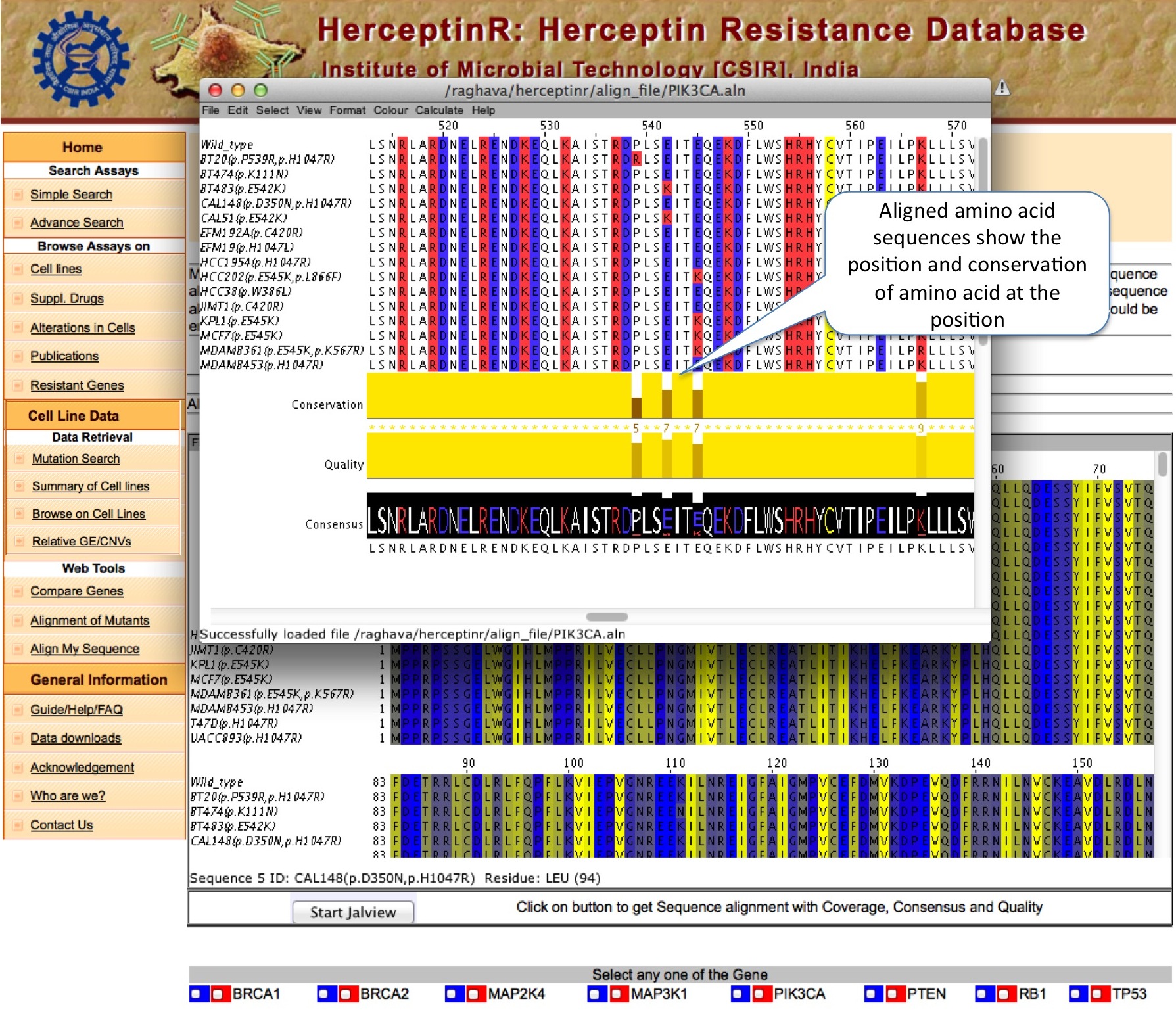
|
|
ALIGN MY SEQUENCE
This module allows user to align their sequence (either protein or gene) with the mutants of important genes to locate the mutation position in their own sequences , if any. User can launch the Jalview by clicking on Start Jalview, which pop-up separate window to show alignment.
|
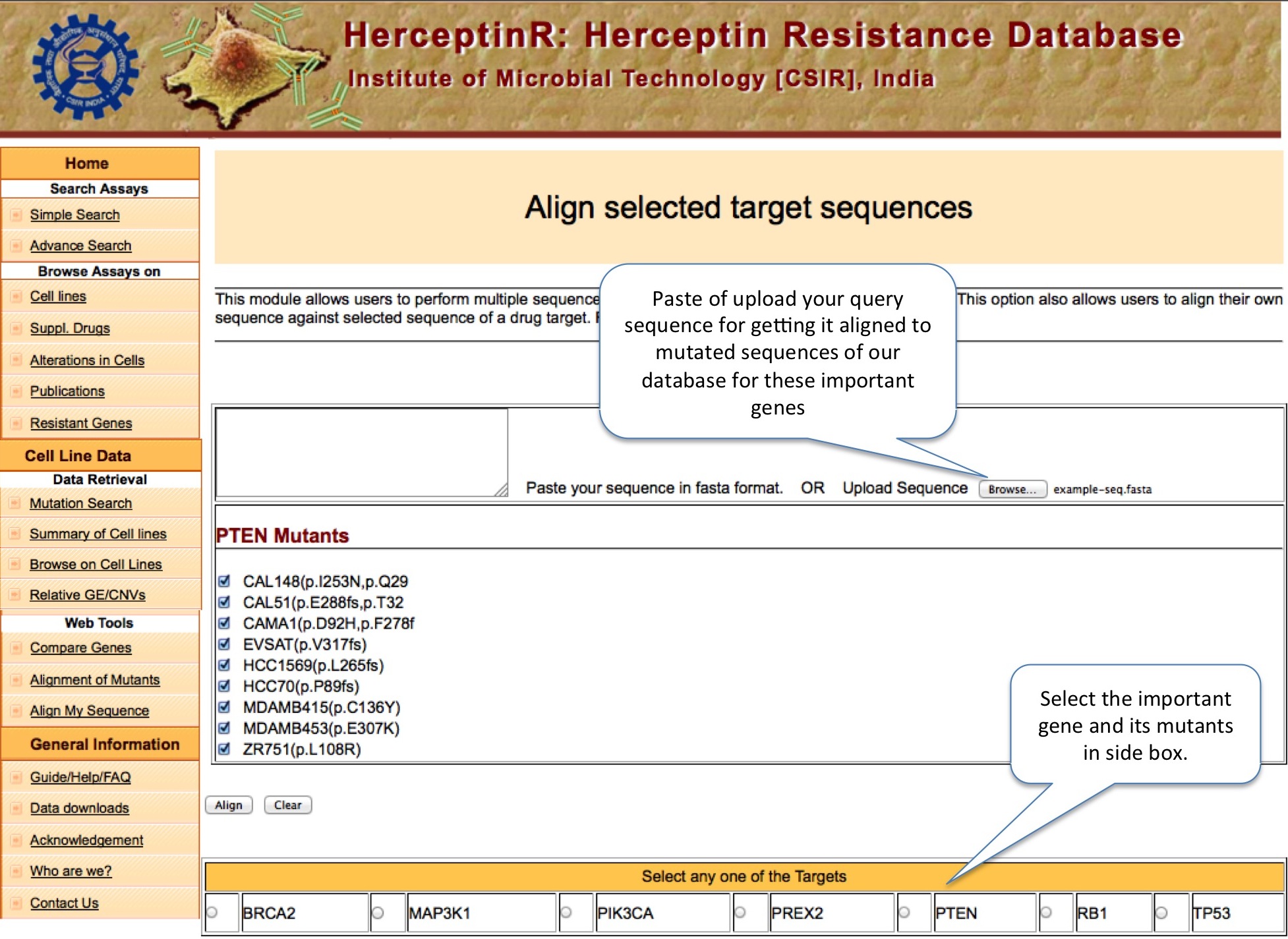
|
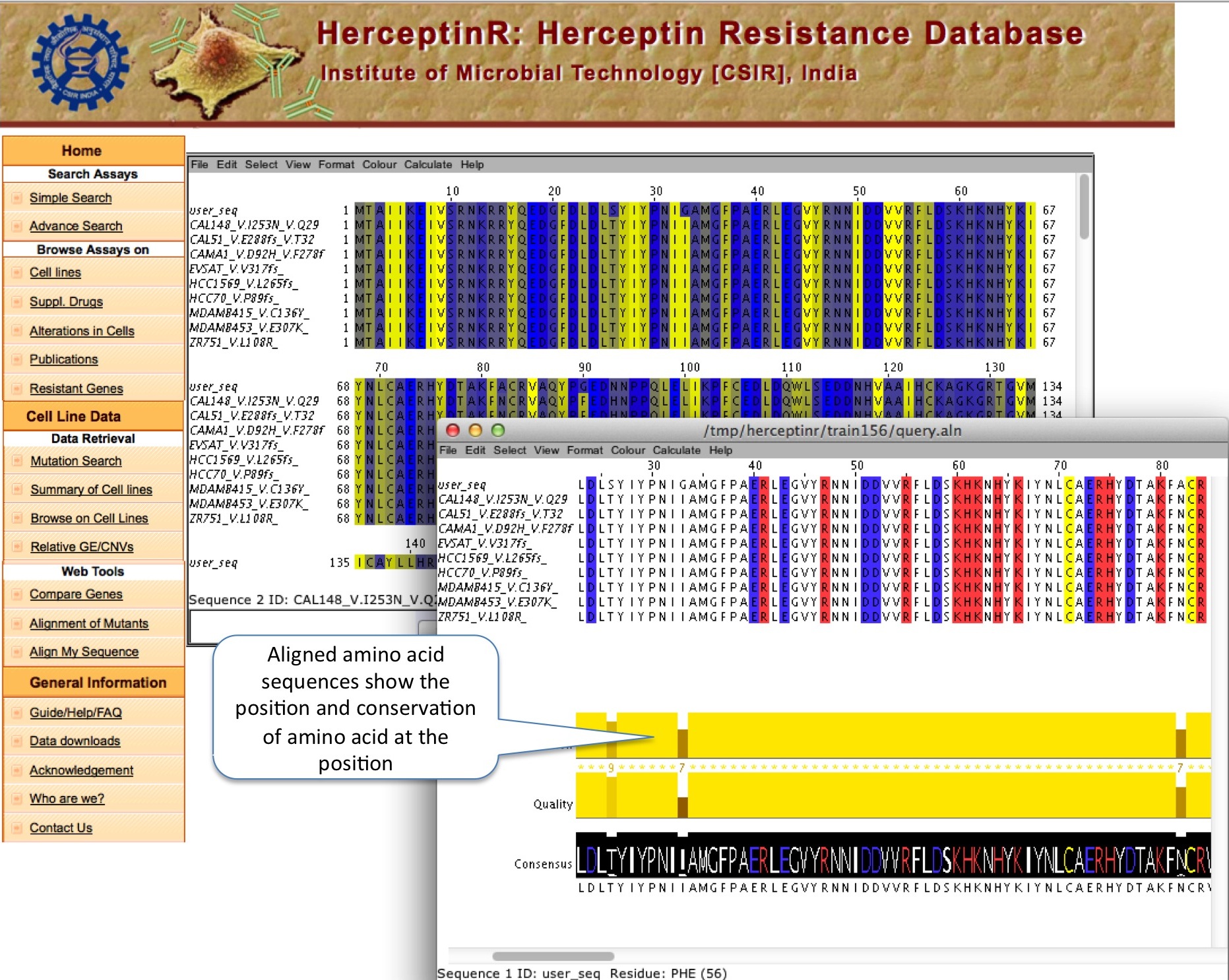
|
|