Introduction to Microbiome
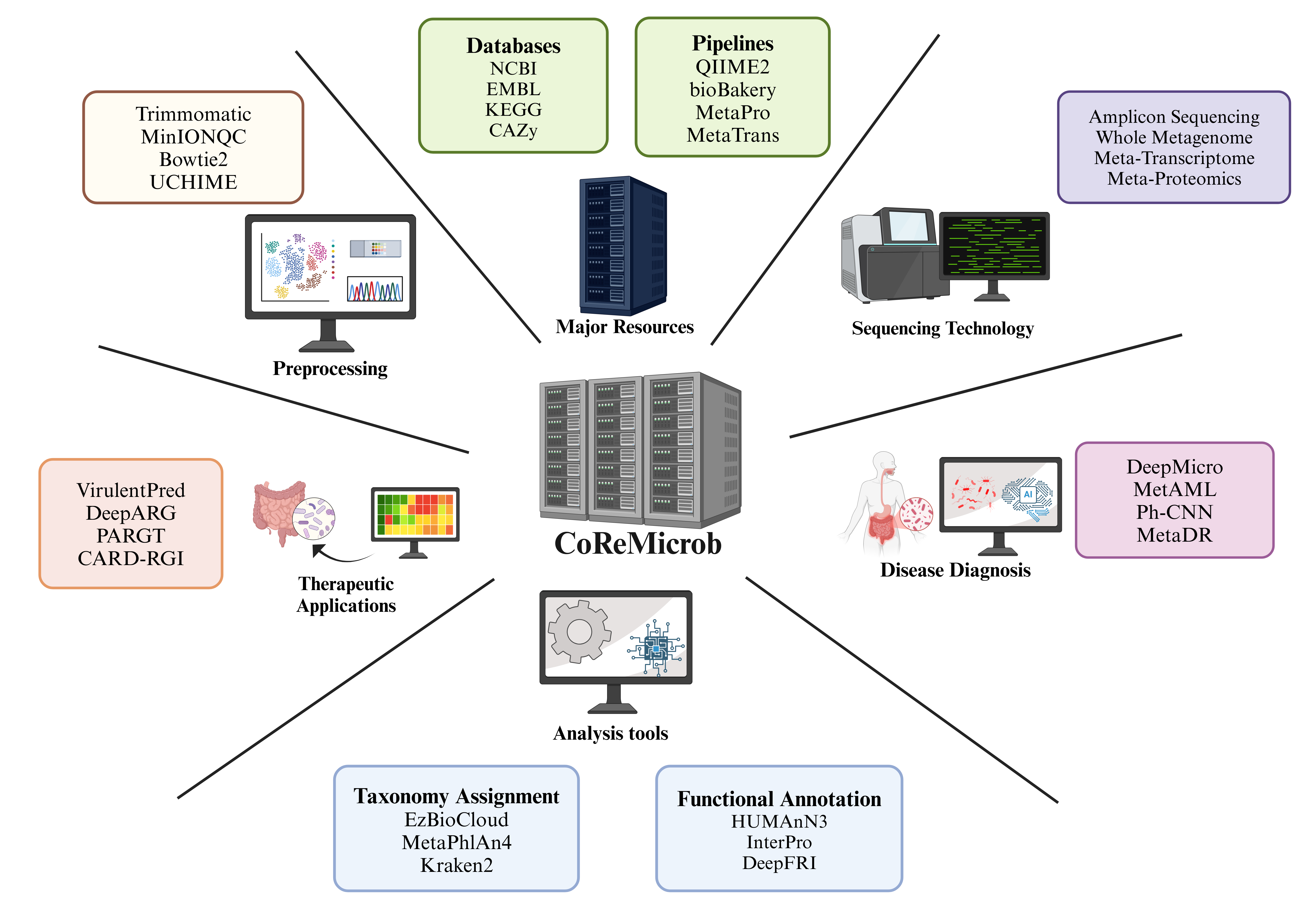
Primary goal of this website is to provide a centralized and comprehensive resource hub for researchers, bioinformaticians, and microbiome scientists working in metagenome analysis. Given the vast and rapidly evolving landscape of metagenomic tools, databases, and software, it can be challenging to identify the most relevant and up-to-date resources for specific analytical needs. By compiling and organizing these resources in one accessible platform, we aim to facilitate efficient exploration, comparison, and utilization of available tools for microbial community analysis, functional annotation, taxonomic profiling, and multi-omics integration. This initiative is designed to enhance research productivity, promote reproducibility, and support the global scientific community in advancing microbiome research and its health, environmental science, and biotechnology applications.
The microbiome refers to the collective genomes of the microorganisms—comprising bacteria, viruses, fungi, and other microbes—that inhabit various environments, including the human body. The microbial communities play crucial roles in maintaining ecological balance and influencing host health.
Metagomics is a powerful tool used to study these complex microbial ecosystems. By analyzing genetic material recovered directly from environmental samples, metagenomics allows researchers to identify and characterize the diversity and functions of microorganisms without the need for culturing them in the laboratory. This approach has been instrumental in uncovering the vast array of microbial life and their potential roles in various environments.
In the case of the human gut, metagenomic studies have revealed that the gut microbiome consists of more than 1,000 microbial species. These microbe are integral to numerous physiological processes, including protection against pathogens, modulation of the immune system, and regulation of metabolic functions. Advancements in sequencing technologies have enabled detailed analyses of these communities, providing insights into their composition and functional capabilities.
Furthermore, metagenomics has facilitated the identification of novel genes, microbial pathways, and antibiotic resistance genes. It has also enhanced our understanding of the interactions co-evolution between microbiota and their hosts. Despite these advancements, challenges remain, such as the need for improved methods to analyze and interpret the vast amounts of data generated. Integrating metagenomics with other 'omics' approaches, like metatranscriptomics, metaproteomics, and metabolomics, holds promise for a more comprehensive understanding of microbial commities and their importance on health and disease.
In summary, metagenomics has revolutionized our understanding of the microbiome, providing a window of the previously hidden world of microbial life and its profound influence on various ecosystems and host organisms.
URL for references used
- Human Microbiome Project Consortium. (2012). Structure, function, and diversity of the healthy human microbiome. Nature, 486(7402), 207–214. https://doi.org/10.1038/nature11234
- Gilbert, J. A., Blaser, M. J., Caporaso, J. G., Jansson, J. K., Lynch, S. V., & Knight, R. (2018). Current understanding of the human microbiome. Nature Medicine, 24(4), 392–400. https://doi.org/10.1038/nm.4517
- Qin, J., Li, R., Raes, J., Arumugam, M., Burgdorf, K. S., Manichanh, C., Nielsen, T., Pons, N., Levenez, F., Yamada, T., Mende, D. R., Li, J., Xu, J., Li, S., Li, D., Cao, J., Wang, B., Liang, H., Zheng, H., Xie, Y., Tap, J., Lepage, P., Bertalan, M., Batto, J. M., Hansen, T., Le Paslier, D., Linneberg, A., Nielsen, H. B., Pelletier, E., Renault, P., Sicheritz-Pontén, T., Turner, K., Zhu, H., Yu, C., Li, S., Jian, M., Zhou, Y., Li, Y., Zhang, X., Li, S., Qin, N., Yang, H., Wang, J., Brunak, S., Doré, J., Guarner, F., Kristiansen, K., Pedersen, O., Parkhill, J., Weissenbach, J., Bork, P., Ehrlich, S. D., & Wang, J. (2010). A human gut microbial gene catalogue established by metagenomic sequencing. Nature, 464(7285), 59–65. https://doi.org/10.1038/nature08821
- Marchesi, J. R., Adams, D. H., Fava, F., Hermes, G. D. A., Hirschfield, G. M., Hold, G., Quraishi, M. N., Kinross, J., Smidt, H., Tuohy, K. M., Thomas, L. V., Zoetendal, E. G., & Shanahan, F. (2016). The gut microbiota and host health: A new clinical frontier. Gut, 65(2), 330–339. https://doi.org/10.1136/gutjnl-2015-309990
- Lloyd-Price, J., Abu-Ali, G., & Huttenhower, C. (2016). The healthy human microbiome. Genome Medicine, 8, 51. https://doi.org/10.1186/s13073-016-0307-y