Cancertope
In silico Platform for designing genome-based
Personalized immunotherapy or Vaccine against Cancer
Personalized immunotherapy or Vaccine against Cancer
Generalised Epitopes |
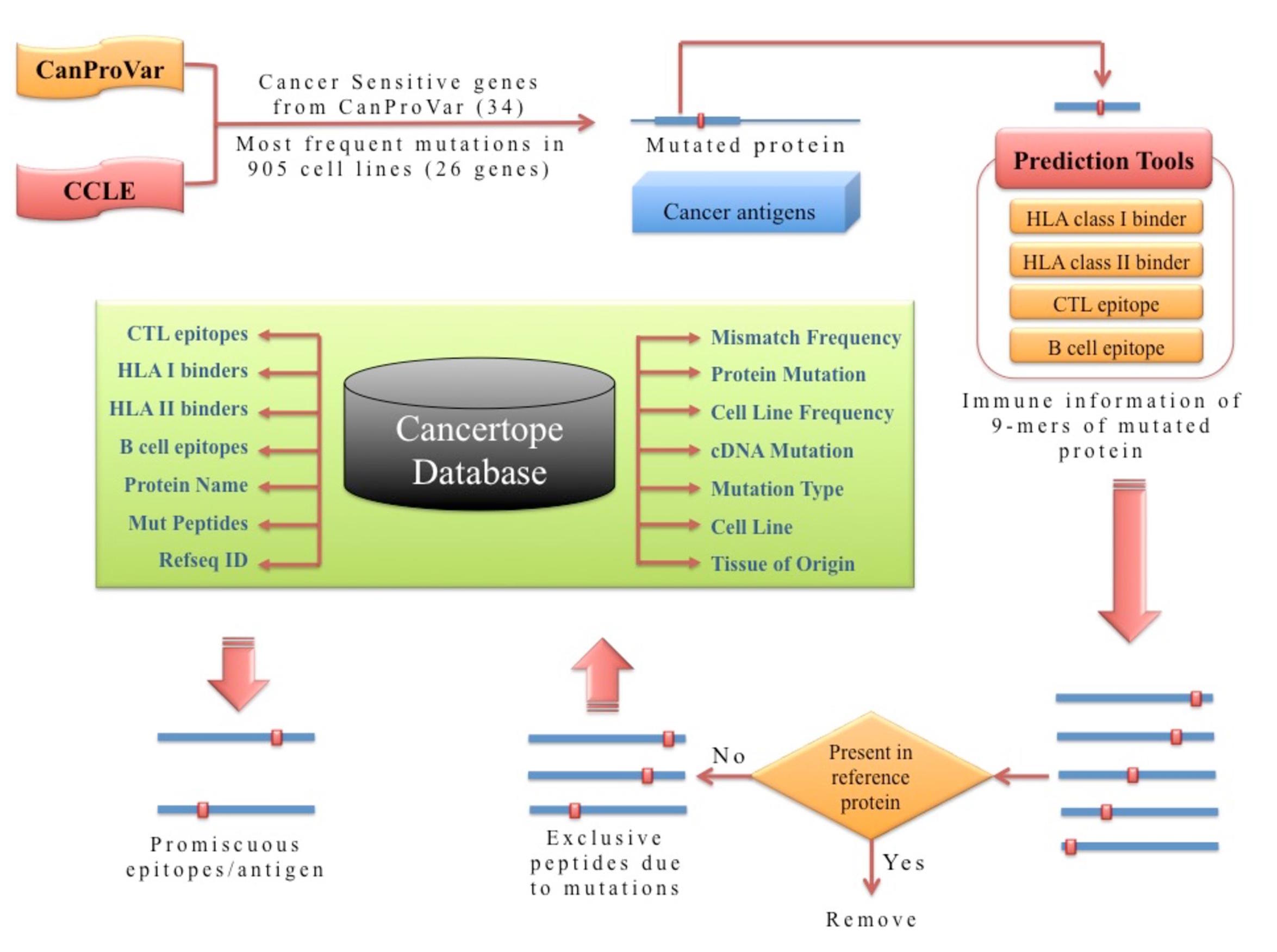
| This module houses neopeptides/neoepitopes generated due to mutation in 60 vaccine candidate over large number of cancer cell lines (905). allows user to explore neopeptides, the tissue of origin, position of neopeptides in protein sequence, the mismatch generated after aligning mutated and reference sequence, the coverage of cell lines with this peptide, CTL epitopes, B cell epitopes and number of MHC alleles (MHC I, MHC II) to which the peptides is predicted to bind. For example, if our query to search all the peptides that were CTL epitopes AND binds with HLA II with “Large Intestine” as their tissue of origin, it will result in the display of a large number of peptides among which ‘FLRHHCPQL’ is a peptide from PDE4DIP protein and present in overall 184 cell lines. Based on these information, one can develop the subunit vaccine or immunotherapy against cancer. |
|
CANCERTOPE | Raghava's Group | IMTECH | CSIR | CRDD | GPSR Package |