Personalized immunotherapy or Vaccine against Cancer
Algorithm |
Data
The mutation data is obtained from CCLE and CanProVar databases.
CanProVar database
We have adopted cancer associated deleterious mutations and general polymorphism from CanProVar database and calculated the ration of frequency of deleterious mutations (fD) over general polymorphism (fP). Such 36 genes showing high ratio (fD/fP) have been selected for mutation and immune epitope analysis..
| Target | UniProt ID | RefSeq ID | Deleterious Mutation Freq (fD) | Polymorphism Freq (fP) | Ratio (fD/fP) | Family |
|---|---|---|---|---|---|---|
| PTEN | P60484 | NM_000314 | 389 | 1 | 389 | NA |
| TP53 | P04637 | NM_001126112 | 1353 | 7 | 193.3 | P53_family |
| CTNNB1 | P35222 | NM_001904 | 132 | 1 | 132 | Beta-catenin_family |
| BRAF | P15056 | NM_004333 | 99 | 1 | 99 | Protein_kinase_superfamily,_TKL_Ser/Thr_protein_kinase_family,_RAF_subfamily |
| NF2 | P35240 | NM_000268 | 74 | 1 | 74 | NA |
| EGFR | P00533 | NM_005228 | 188 | 3 | 62.7 | Protein_kinase_superfamily,_Tyr_protein_kinase_family,_EGF_receptor_subfamily |
| SMAD4 | Q13485 | NM_005359 | 107 | 2 | 53.5 | Dwarfin/SMAD_family |
| SMAD4 | Q13485 | NM_005359 | 107 | 2 | 53.5 | Dwarfin/SMAD_family |
| VHL | P40337 | NM_000551 | 272 | 6 | 45.3 | NA |
| KIT | P10721 | NM_000222 | 131 | 3 | 43.7 | Protein_kinase_superfamily,_Tyr_protein_kinase_family,_CSF-1/PDGF_receptor_subfamily |
| PIK3CA | P42336 | NM_006218 | 174 | 4 | 43.5 | PI3/PI4-kinase_family |
| NRAS | P01111 | NM_002524 | 36 | 1 | 36 | Small_GTPase_superfamily,_Ras_family |
| MSH2 | P43246 | NM_000251 | 103 | 5 | 20.6 | DNA_mismatch_repair_MutS_family |
| GATA1 | P15976 | NM_002049 | 20 | 1 | 20 | NA |
| MLH1 | P40692 | NM_000249 | 118 | 6 | 19.7 | DNA_mismatch_repair_MutL/HexB_family |
| FBXW7 | Q969H0 | NM_033632 | 67 | 4 | 16.8 | NA |
| MEN1 | O00255 | NM_130800 | 49 | 3 | 16.3 | NA |
| FGFR3 | P22607 | NM_001163213 | 31 | 2 | 15.5 | Protein_kinase_superfamily,_Tyr_protein_kinase_family,_Fibroblast_growth_factor_receptor_subfamily |
| TSHR | P16473 | NM_000369 | 46 | 3 | 15.3 | G-protein_coupled_receptor_1_family,_FSH/LSH/TSH_subfamily |
| JAK2 | O60674 | NM_004972 | 40 | 3 | 13.3 | Protein_kinase_superfamily,_Tyr_protein_kinase_family,_JAK_subfamily |
| RB1 | P06400 | NM_000321 | 102 | 8 | 12.8 | Retinoblastoma_protein_(RB)_family |
| PDGFRA | P16234 | NM_006206 | 35 | 3 | 11.7 | Protein_kinase_superfamily,_Tyr_protein_kinase_family,_CSF-1/PDGF_receptor_subfamily |
| NF1 | P21359 | NM_001042492 | 65 | 6 | 10.8 | NA |
| FGFR2 | P21802 | NM_022970 | 43 | 4 | 10.8 | Protein_kinase_superfamily,_Tyr_protein_kinase_family,_Fibroblast_growth_factor_receptor_subfamily |
| FLT3 | P36888 | NM_004119 | 35 | 4 | 8.8 | Protein_kinase_superfamily,_Tyr_protein_kinase_family,_CSF-1/PDGF_receptor_subfamily |
| CDH1 | P12830 | NM_004360 | 68 | 8 | 8.5 | NA |
| TNFAIP3 | P21580 | NM_006290 | 31 | 4 | 7.8 | Peptidase_C64_family |
| CBL | P22681 | NM_005188 | 30 | 4 | 7.5 | NA |
| RET | P07949 | NM_020975 | 58 | 8 | 7.3 | Protein_kinase_superfamily,_Tyr_protein_kinase_family |
| MSH6 | P52701 | NM_000179 | 40 | 8 | 5 | DNA_mismatch_repair_MutS_family |
| ERBB2 | P04626 | NM_004448 | 29 | 6 | 4.8 | Protein_kinase_superfamily,_Tyr_protein_kinase_family,_EGF_receptor_subfamily |
| MET | P08581 | NM_001127500 | 23 | 5 | 4.6 | Protein_kinase_superfamily,_Tyr_protein_kinase_family |
| ABL1 | P00519 | NM_007313 | 23 | 7 | 3.3 | Protein_kinase_superfamily,_Tyr_protein_kinase_family,_ABL_subfamily |
| ALK | Q9UM73 | NM_004304 | 27 | 9 | 3 | Protein_kinase_superfamily,_Tyr_protein_kinase_family,_Insulin_receptor_subfamily |
| ATM | Q13315 | NM_000051 | 134 | 52 | 2.6 | PI3/PI4-kinase_family,_ATM_subfamily |
CCLE database
In other approach we looked at most frequent mutations in the CCLE database. In this study we looked at those mutations which were found in at least 10% of total cell lines (905)..
| Target | UniProt ID | RefSeq ID | Family |
|---|---|---|---|
| AAK1 | Q2M2I8 | NM_014911 | Protein kinase superfamily, Ser/Thr protein kinase family |
| AKAP12 | Q02952 | NM_005100 | NA |
| AKAP12 | Q86TJ9 | NM_005100 | NA |
| AKAP9 | Q99996 | NM_005751 | NA |
| AKAP9 | Q5GIA7 | NM_005751 | NA |
| AKAP9 | Q6PJH3 | NM_005751 | NA |
| ALPK2 | Q86TB3 | NM_052947 | Protein kinase superfamily, Alpha-type protein kinase family, ALPK subfamily |
| CARD10 | Q9BWT7 | NM_014550 | NA |
| CHD1 | B3KT33 | NM_001270 | NA |
| CHD1 | O14646 | NM_001270 | SNF2/RAD54 helicase family |
| CREB3L2 | Q68D60 | NM_194071 | NA |
| CREB3L2 | Q70SY1 | NM_194071 | BZIP family, ATF subfamily |
| CTBP2 | P56545 | NM_022802 | D-isomer specific 2-hydroxyacid dehydrogenase family |
| FMN2 | Q9NZ56 | NM_020066 | Formin homology family, Cappuccino subfamily |
| FMN2 | Q9HBL1 | NM_020066 | NA |
| GPR112 | Q8IZF6 | NM_153834 | G-protein coupled receptor 2 family, LN-TM7 subfamily |
| HSP90B1 | V9HWP2 | NM_003299 | NA |
| HSP90B1 | P14625 | NM_003299 | Heat shock protein 90 family |
| MAML2 | Q8IZL2 | NM_032427 | Mastermind family |
| MAP3K1 | Q13233 | NM_005921 | Protein kinase superfamily, STE Ser/Thr protein kinase family, MAP kinase kinase kinase subfamily |
| MAP3K4 | Q9P1M2 | NM_005922 | NA |
| MAP3K4 | Q9Y6R4 | NM_005922 | Protein kinase superfamily, STE Ser/Thr protein kinase family, MAP kinase kinase kinase subfamily |
| MLL3 | Q8NEZ4 | NM_170606 | Class V-like SAM-binding methyltransferase superfamily, Histone-lysine methyltransferase family, TRX/MLL subfamily |
| MSH3 | P20585 | NM_002439 | DNA mismatch repair MutS family, MSH3 subfamily |
| MYLK | Q15746 | NM_053025 | Protein kinase superfamily, CAMK Ser/Thr protein kinase family |
| MYST4 | Q8WYB5 | NM_012330 | MYST (SAS/MOZ) family |
| MYST4 | B2RWN8 | NM_012330 | NA |
| NCOA3 | Q9Y6Q9 | NM_181659 | SRC/p160 nuclear receptor coactivator family |
| NR1H2 | P55055 | NM_007121 | Nuclear hormone receptor family, NR1 subfamily |
| NR1H2 | F1D8P7 | NM_007121 | Nuclear hormone receptor family |
| PDE4DIP | Q5VU43 | NM_014644 | NA |
| PIK3C2G | B7ZLY6 | NM_004570 | NA |
| PIK3C2G | O75747 | NM_004570 | PI3/PI4-kinase family |
| PRKDC | P78527 | NM_006904 | PI3/PI4-kinase family |
| RECQL4 | O94761 | NM_004260 | Helicase family, RecQ subfamily |
| TNRC6B | Q9UPQ9 | NM_001162501 | GW182 family |
| TTBK1 | Q5TCY1 | NM_032538 | Protein kinase superfamily, CK1 Ser/Thr protein kinase family |
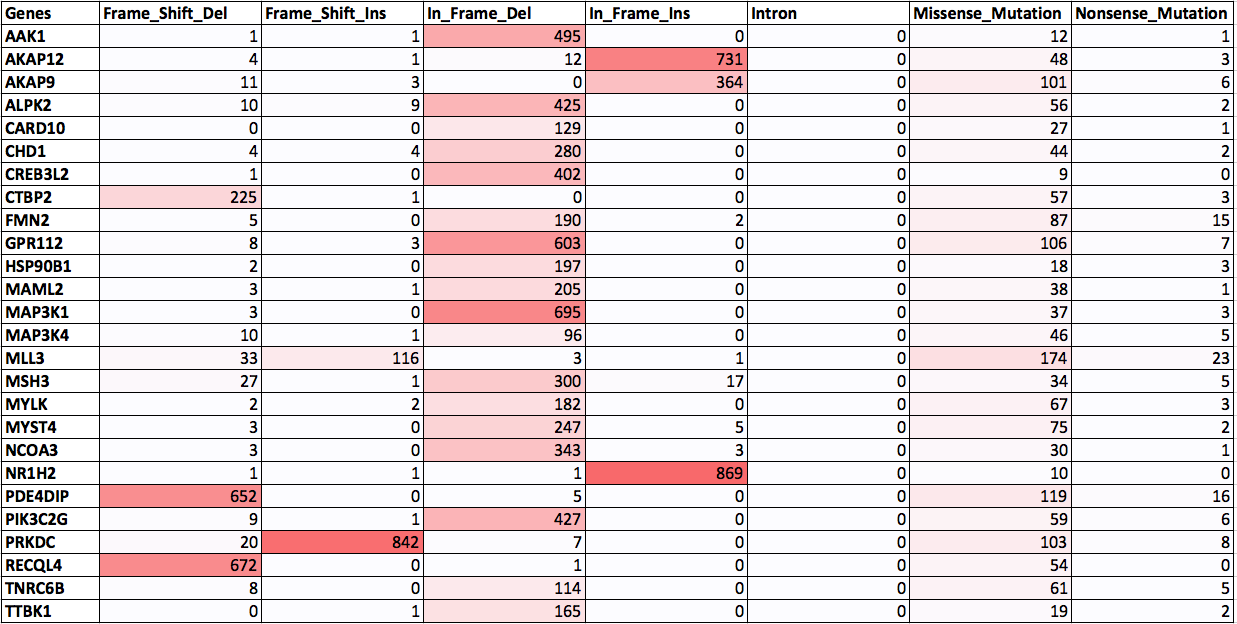
Distribution of frequently mutated genes in CCLE database:
Prediction Algorithms
CTLPred Link
CTLPred is a direct method for prediction of CTL epitopes crucial in subunit vaccine design.In direct methods the information or patterns of T cell epitopes instead of MHC binders were used for the development o f methods. The methods is based on elegant machine learning techniques like Artificial Neural network.
ProPred1 Link
The ProPred-I is an on-line service for identifying the MHC Class-I binding regions in antigens. It implements matrices for 47 MHC Class-I alleles, proteasomal and immunoproteasomal models. The main aim of this server is to help users in identifying the promiscuous regions.
ProPred Link
The aim of this server is to predict MHC Class-II binding regions in an antigen sequence, using quantitative matrices derived from published literature by Sturniolo et. al., 1999. The server will assist in locating promiscuous binding regions that are useful in selecting vaccine candidates.
LBTope Link
The aim of LBTope server is to predict B cell epitope(s) in an antigen sequence, using Support Vector Machine as machine learning method.
|
CANCERTOPE | Raghava's Group | IMTECH | CSIR | CRDD | GPSR Package |