Cancertope
In silico Platform for designing genome-based
Personalized immunotherapy or Vaccine against Cancer
Personalized immunotherapy or Vaccine against Cancer
Guide |
SEARCH |
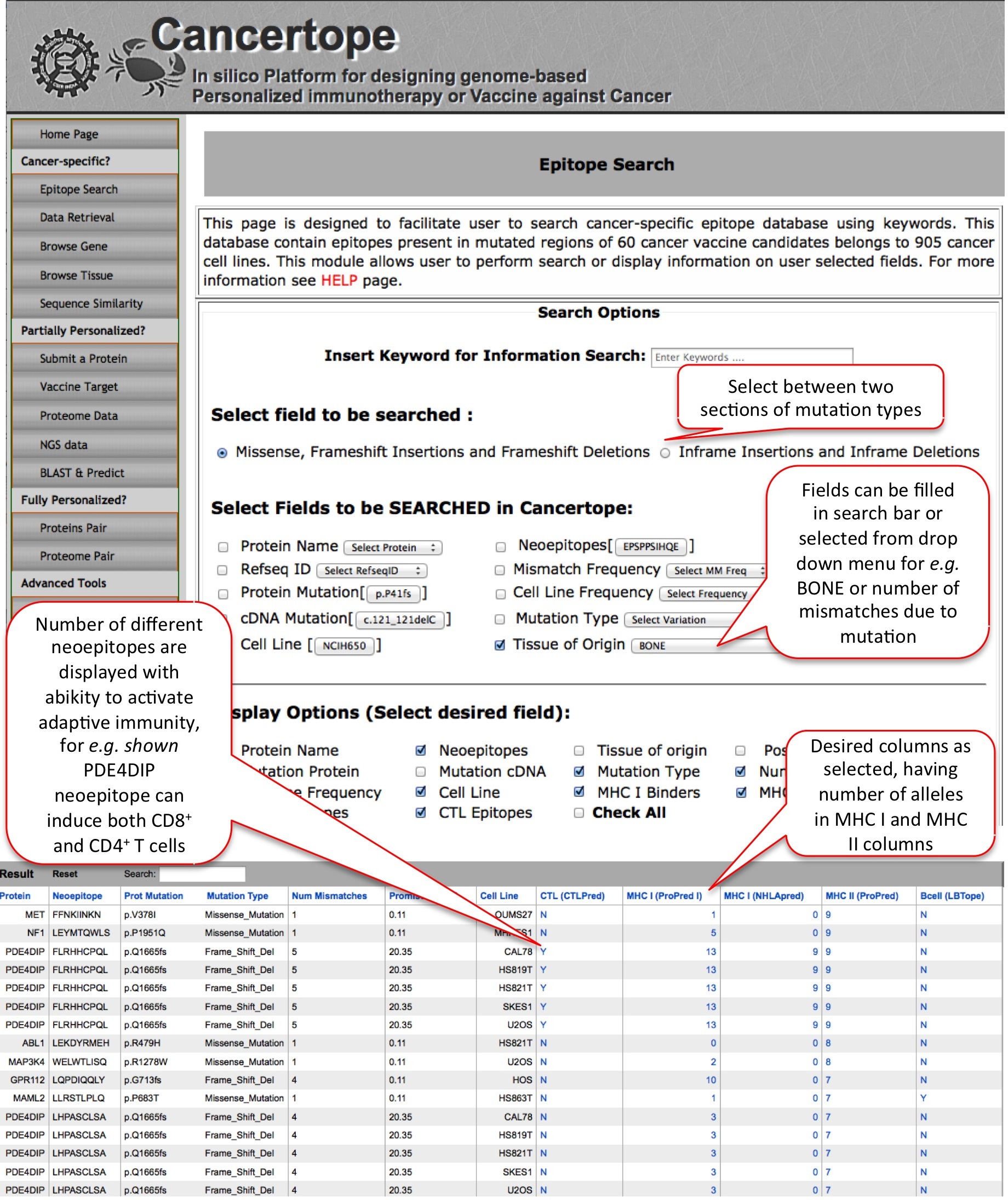
Epitope Search This module deals with the simple search of cancer-specific database section of our study. In this module the user can select or fill his query as shown. for example, here if we want to search all neoepitopes, which are generated due to missense & frame-shift mutation data and found in bone-tissue cell lines, we get number of good candidates. Among several examples below, candidate like FLRHHCPQL from PDE4DIP protein is having 5 mismatches due to frame shift mutation and has capability of inducing CTLs, HLA I and HLA II. In addition this it is very frequent and found in 20.35% of 905 cell lines. |
 |
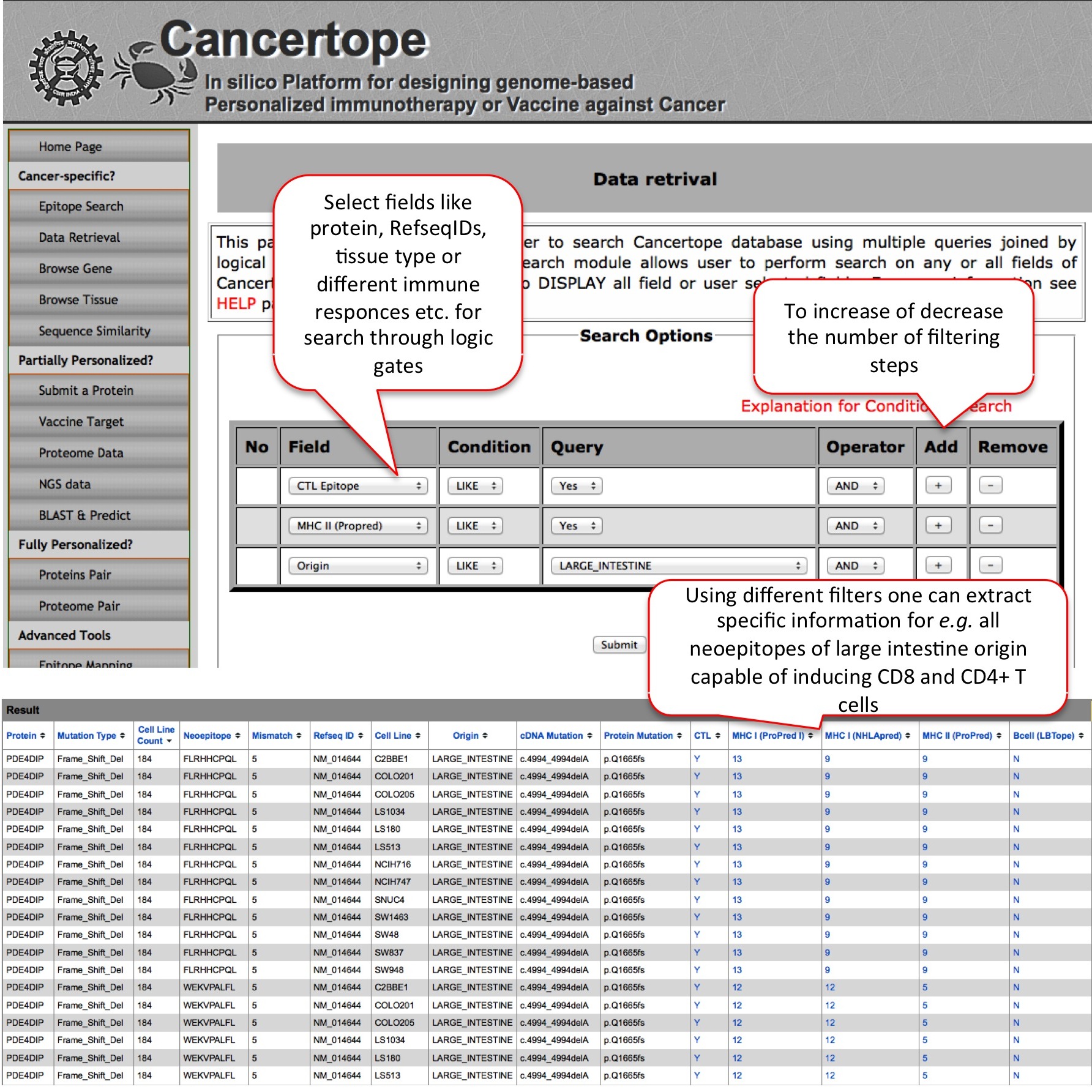
Data Retrival This module is one of the most useful module of our cancer-sepcific database section. Here user can do advanced searching based on logic GATES. For example if user wants to inspect the CTL & CD4+ T cell inducing neoepitopes from large intestine cancer cell lines, the selection of these filters lead to large number of neoepitopes like FLRHHCPQL from PDE4DIP protein. The epitope FLRHHCPQL is having 5 mismatches due to frame shift mutation and has capability of inducing CTLs, HLA I and HLA II. |
 |
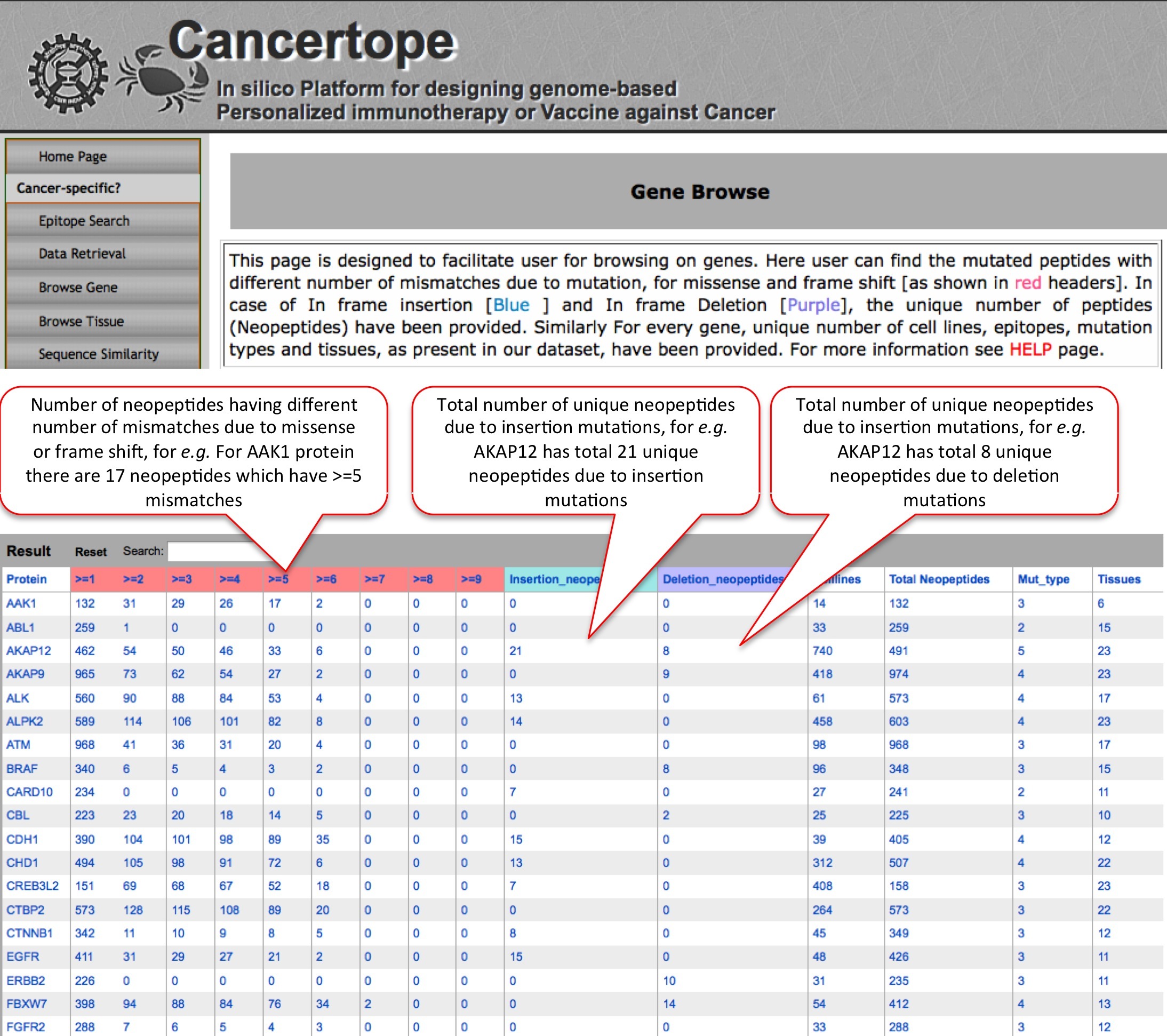
Browse Gene This module of database section allows user to have a overview on complete database with respect to cancer vaccine targets. |
 |
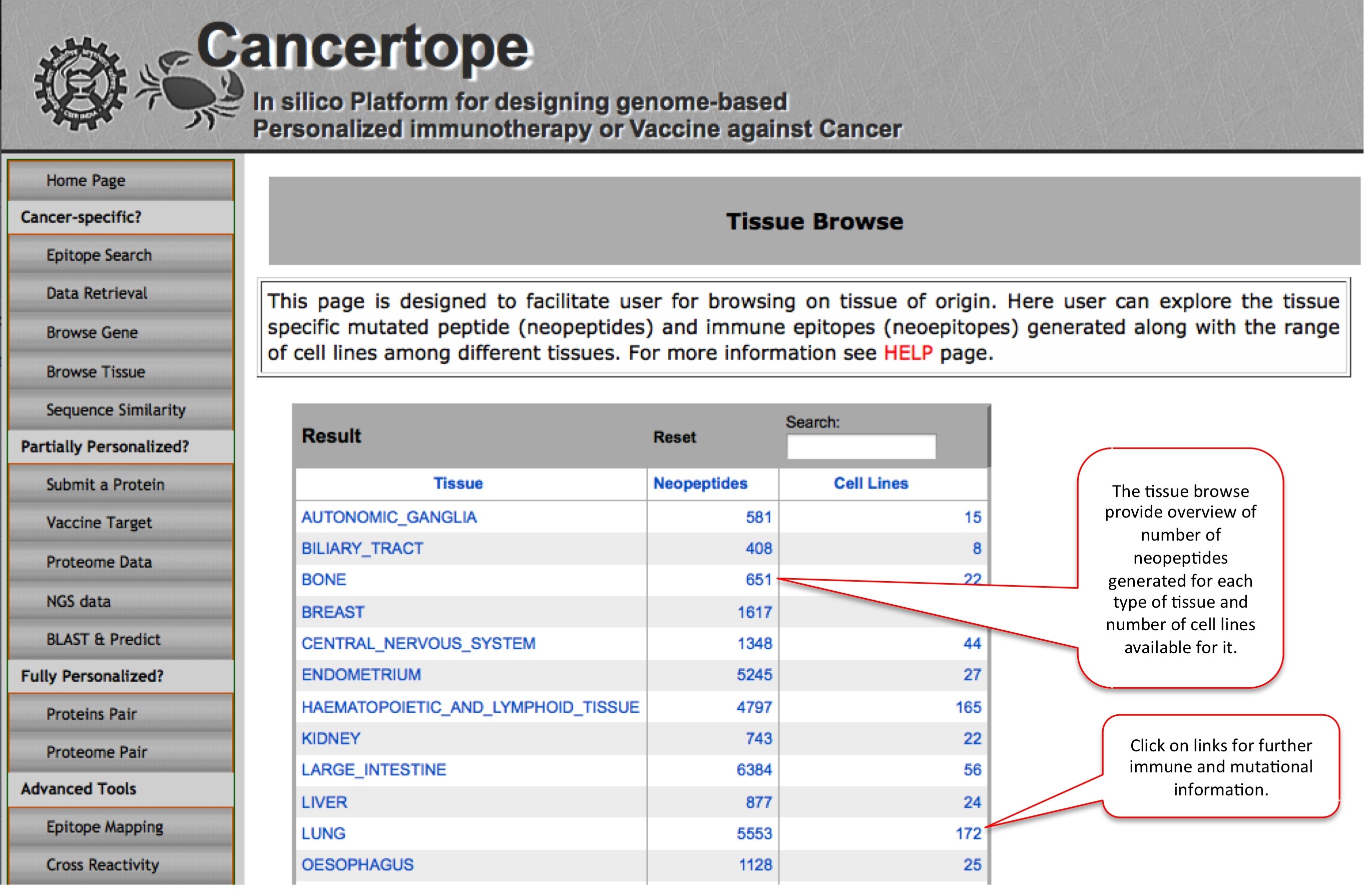
Browse Tissue |
 |
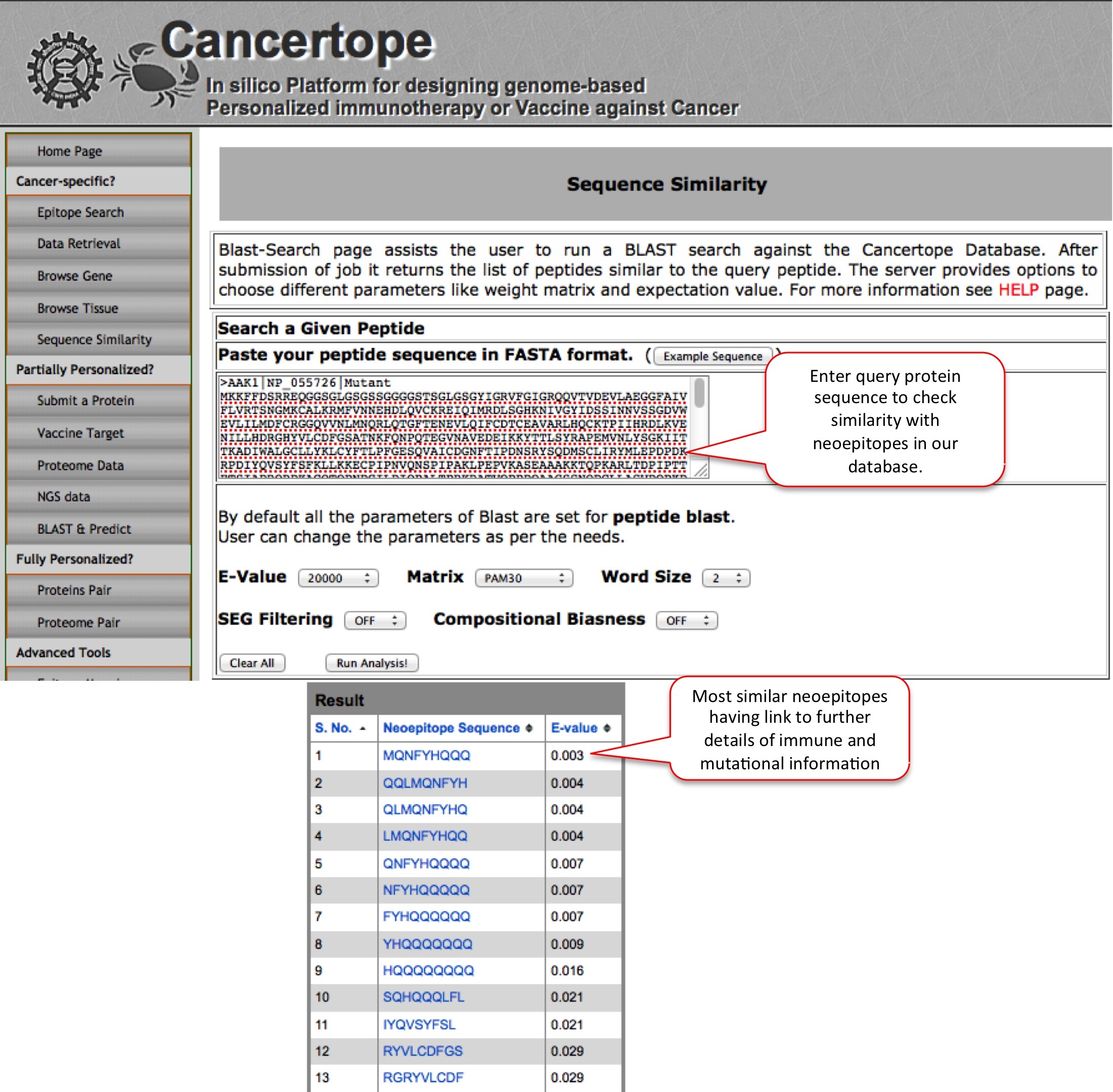
Sequence Similarity In this module the user can do sequence similarity search for his query in the database section of Cancertope. For example the ALK mutant was submitted below to get all the similar neoepitopes that exist in Cancertope database section. This search leads to a very similar neoepitope MQNFYHQQQ with a E Value of 0.003. The epitope like leads to the further details of neoepitope. |
 |
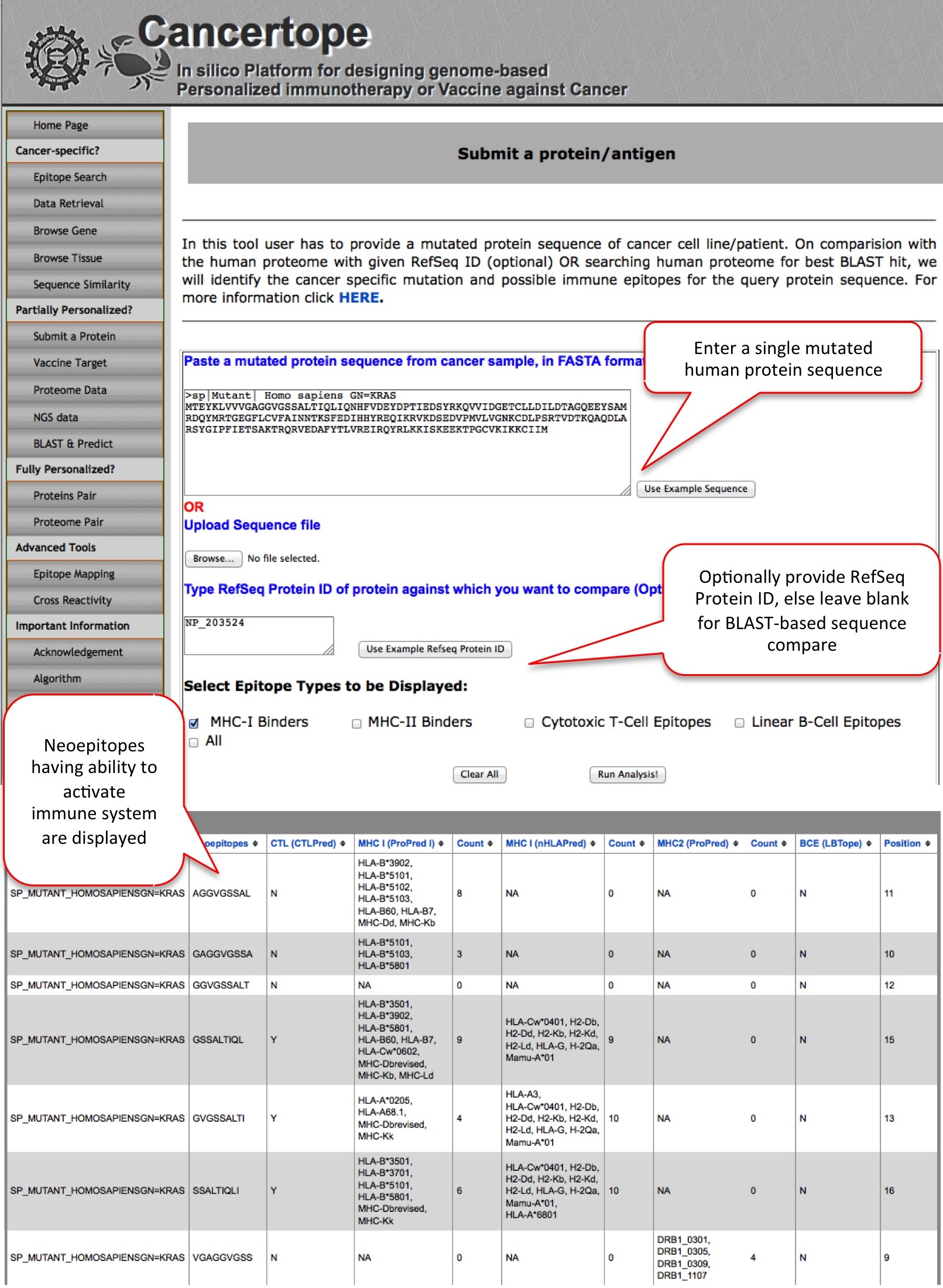
Submit a Protein This module is a part of partially personalized section of Cancertope. Here the user can submit a single mutant protein sequence and user can investigate if there are some neoepitopes (generated due to mutation) in the query which have ability to induce adaptive immunity and have least/no cross reactivity. For example, here user submit a mutant KRAS protein sequence for identification and prediction of neoepitopes. User may or may not provide RefSeq Protein ID. In case the RefSeq Protein ID is not provided, module gets the best BLAST hit from human reference proteome for the query and compares the wild type sequence with mutant sequence. This leads to identification of de novo region (neopeptides) in the query mutant protein. Later the neopeptides are predicted with the pipeline, to get neoepitopes. |
 |
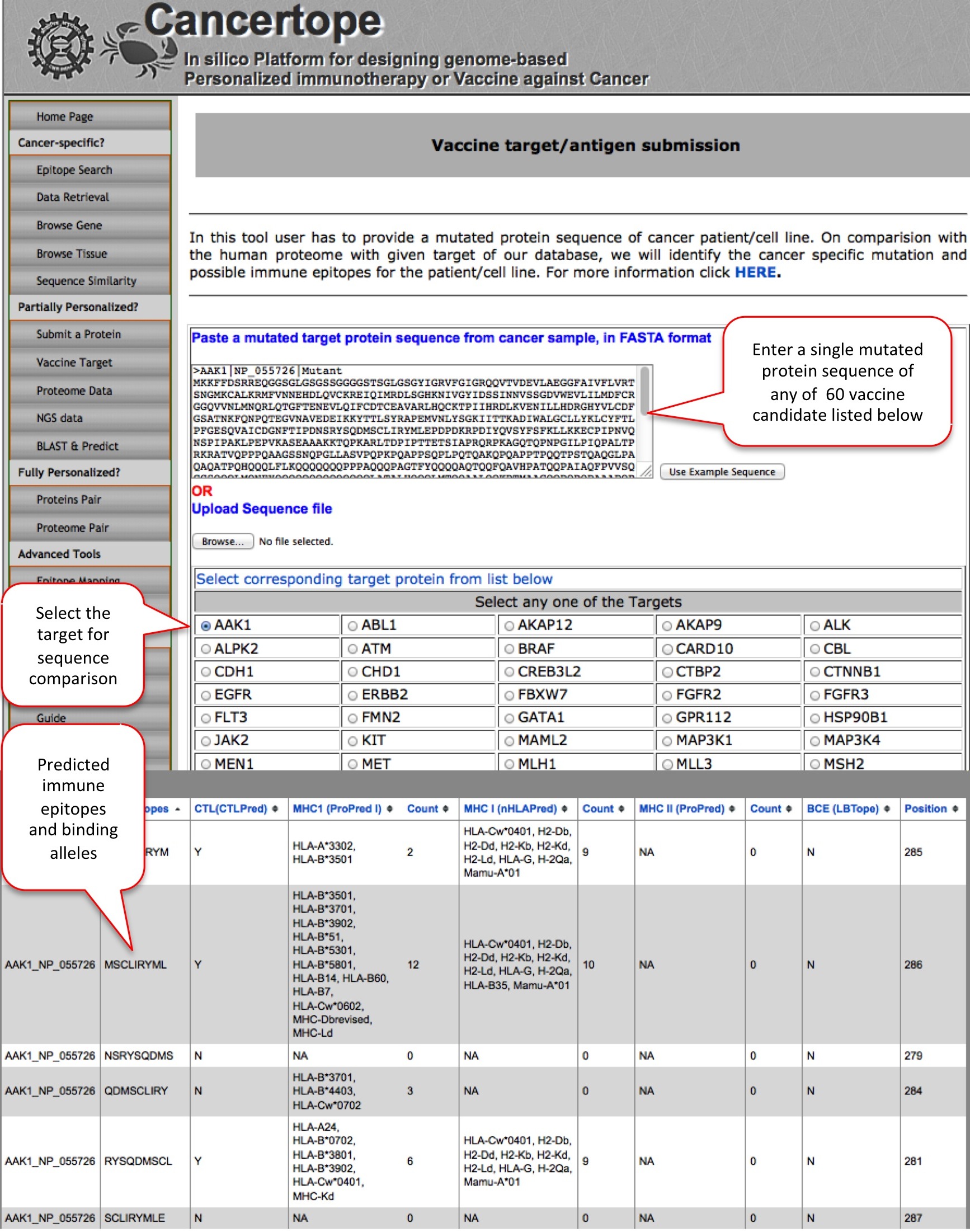
Vaccine Target Similar to the "Submit a protein" module, here the user can submit a mutant of vaccine target sequence, as provided in the list. |
 |
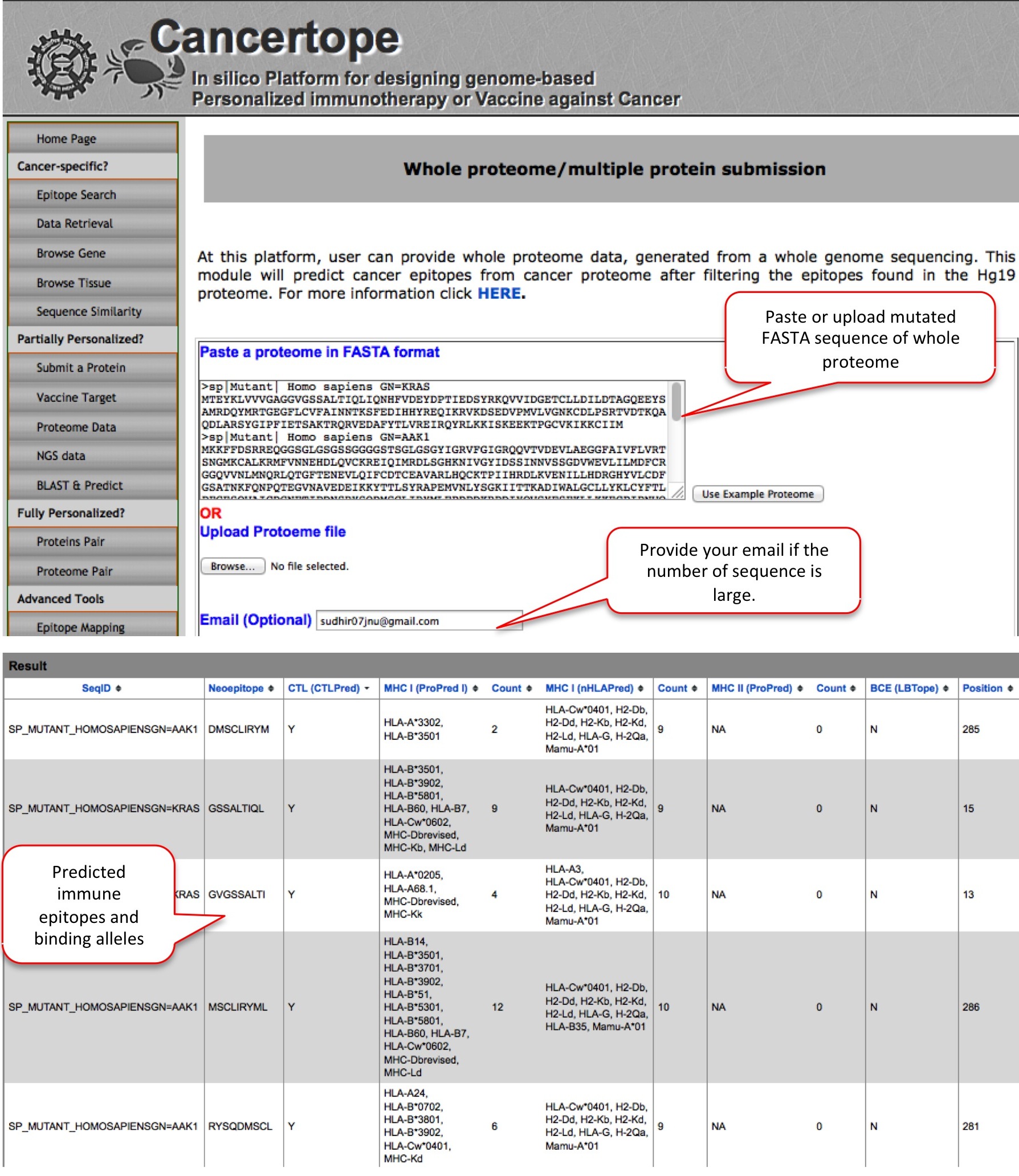
Proteome Data As a part of partially personalized section of Cancertope, here the user can submit multiple mutant protein sequences or whole proteome sequence from cancer sample. On submission, user can investigate the neoepitopes (generated due to mutation) in the query which have ability to induce adaptive immunity and have least/no cross reactivity. |
 |
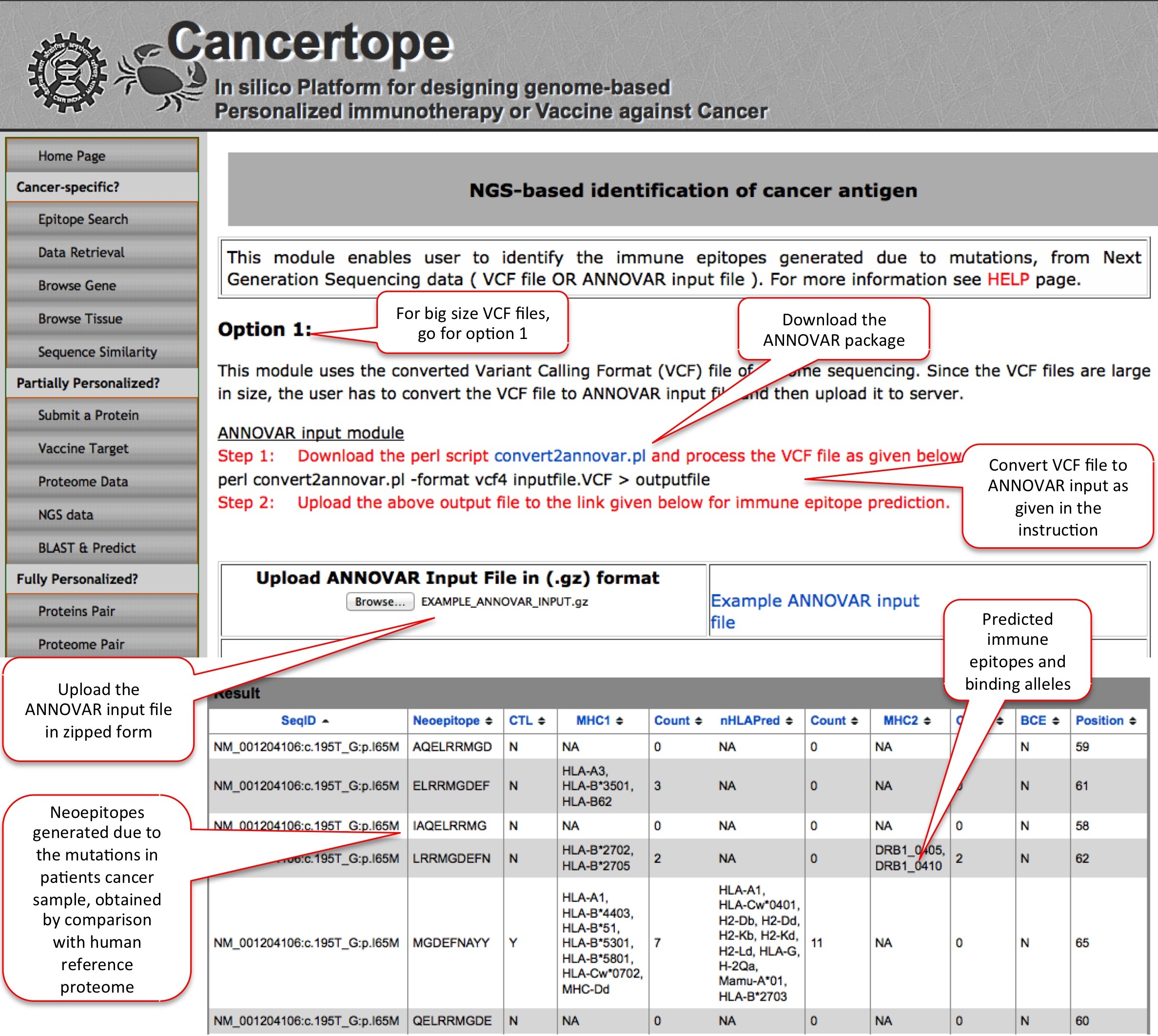
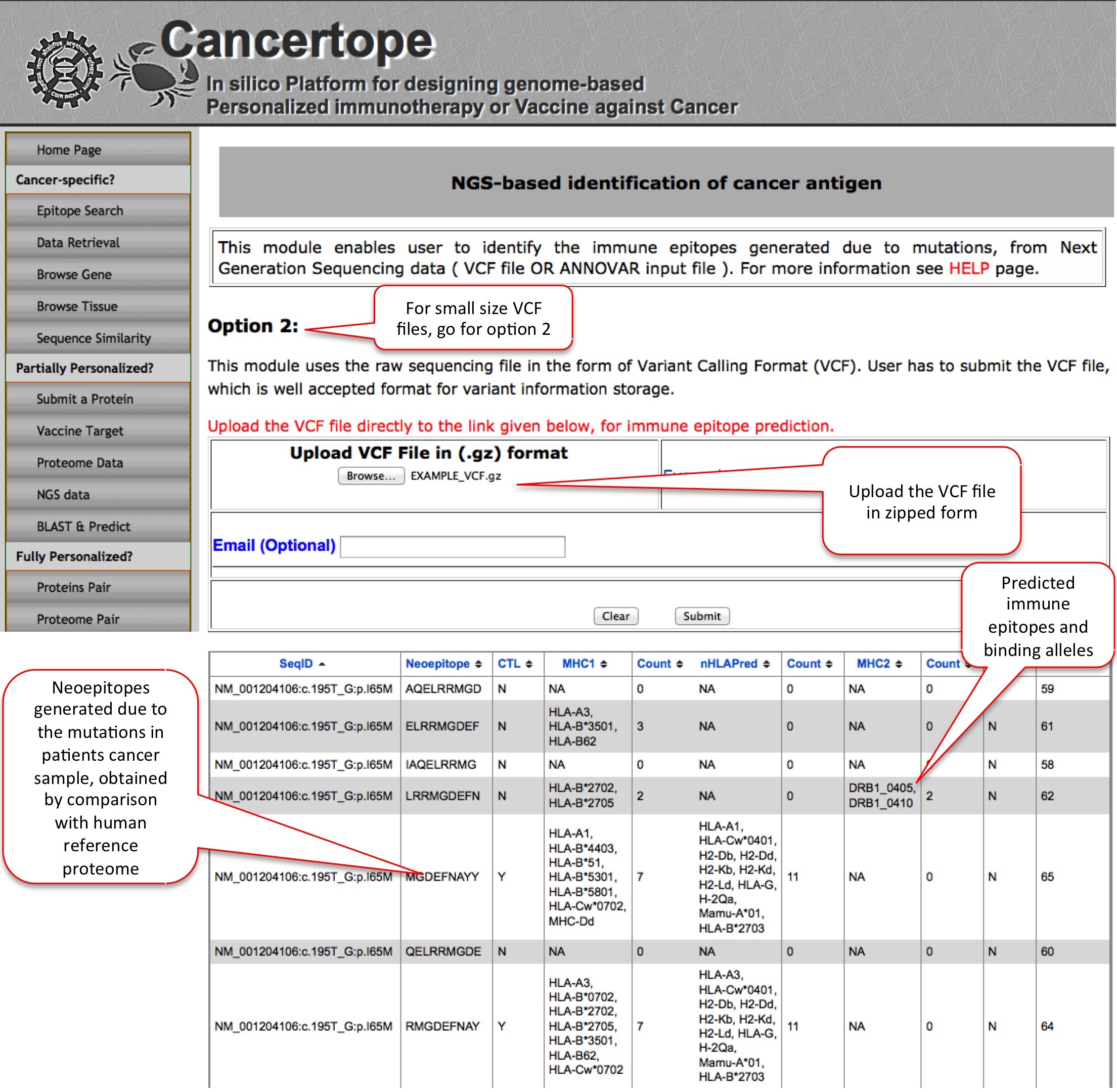
NGS Data This module is one of the most important module of partially personalized section. Since the raw NGS data comes in the VCF format, we designed this module which can annotate the mutated proteins from VCF files, and indentify the de novo region (neopeptides) in the mutant protein sequences. The neopeptides are then further predicted through pipeline to get neoepitopes. In the option 1 the user has to download the a PERL program from ANNOVAR website by which user can reduce the big sized VCF file to small sized ANNOVAR input. In contrast, in Option 2 user can directly submit the VCF file provided the size is not very big. |
 |
 |
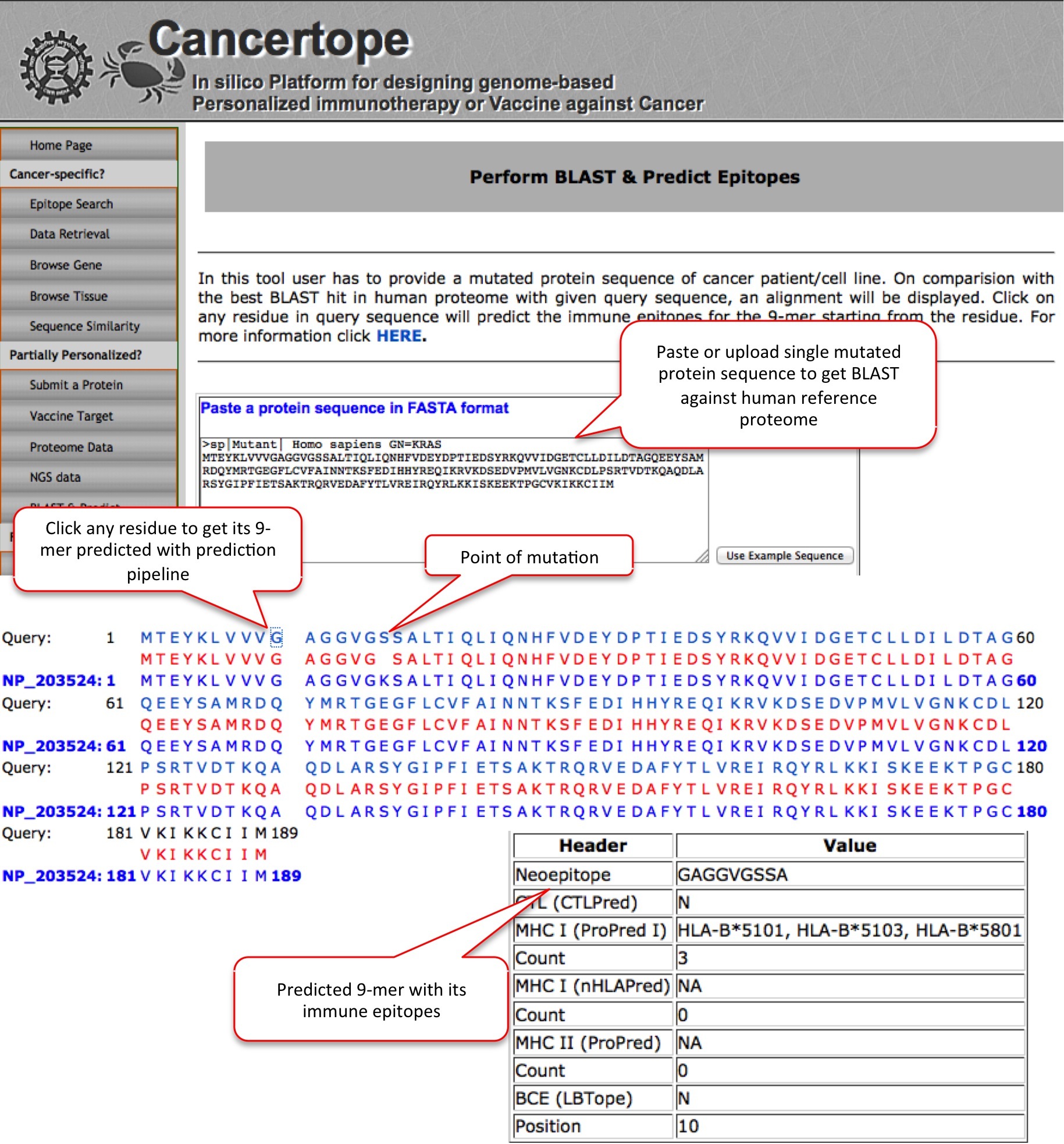
BLAST & Predict This module enables user to BLAST its query against the human reference proteome and the resultant BLAST output is presented where user can selected any residue on query protein to get its 9-mer predicted through pipeline. FOr example a mutant of KRAS is submited here which on BLAST hit gets its wild type sequence in alighment. If we select say the 10th residue, we get prediction on GAGGVGSSA peptide. |
 |
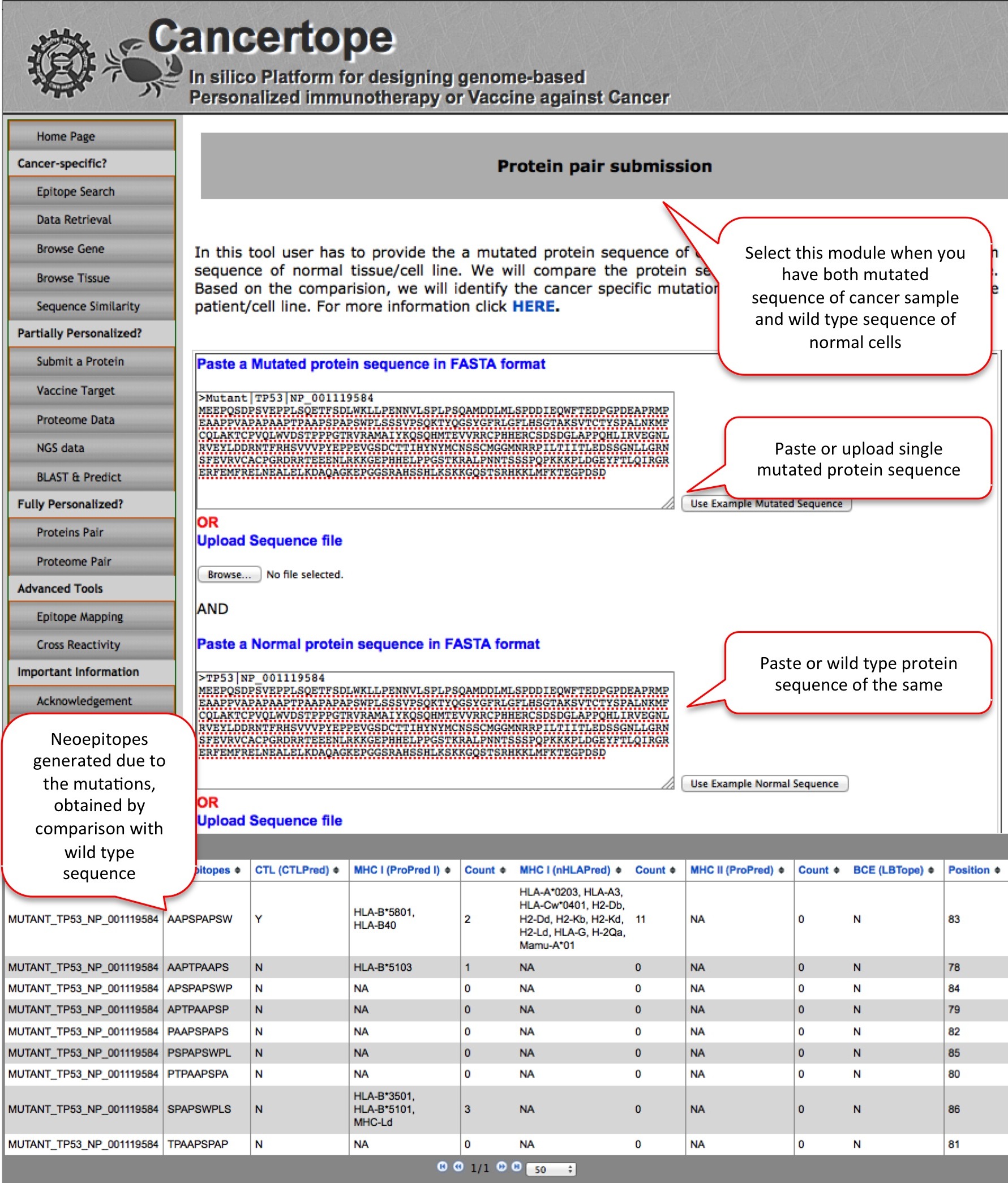
Protein Pair THis module is a part f fully personalized section, where the user need to have both, a protein sequence from cancer sample and sequence of same protein from healthy tissue of same individual. THe module compares the mutant and wild type sequences and identify the region with mutation. THe mutated region is later predicted for the presence of immune epitopes. |
 |
Proteome Pair Similar to the Protein pair module of fully personalized section, the proteome pair module requir the proteome sequence/multiple sequences from cancer sample and proteome or same multiple sequences from healthy sample. The comparison between cancerous and healthy sample sequences lead to identification of mutated region and its peptides which are later predicted for immune epitopes. |
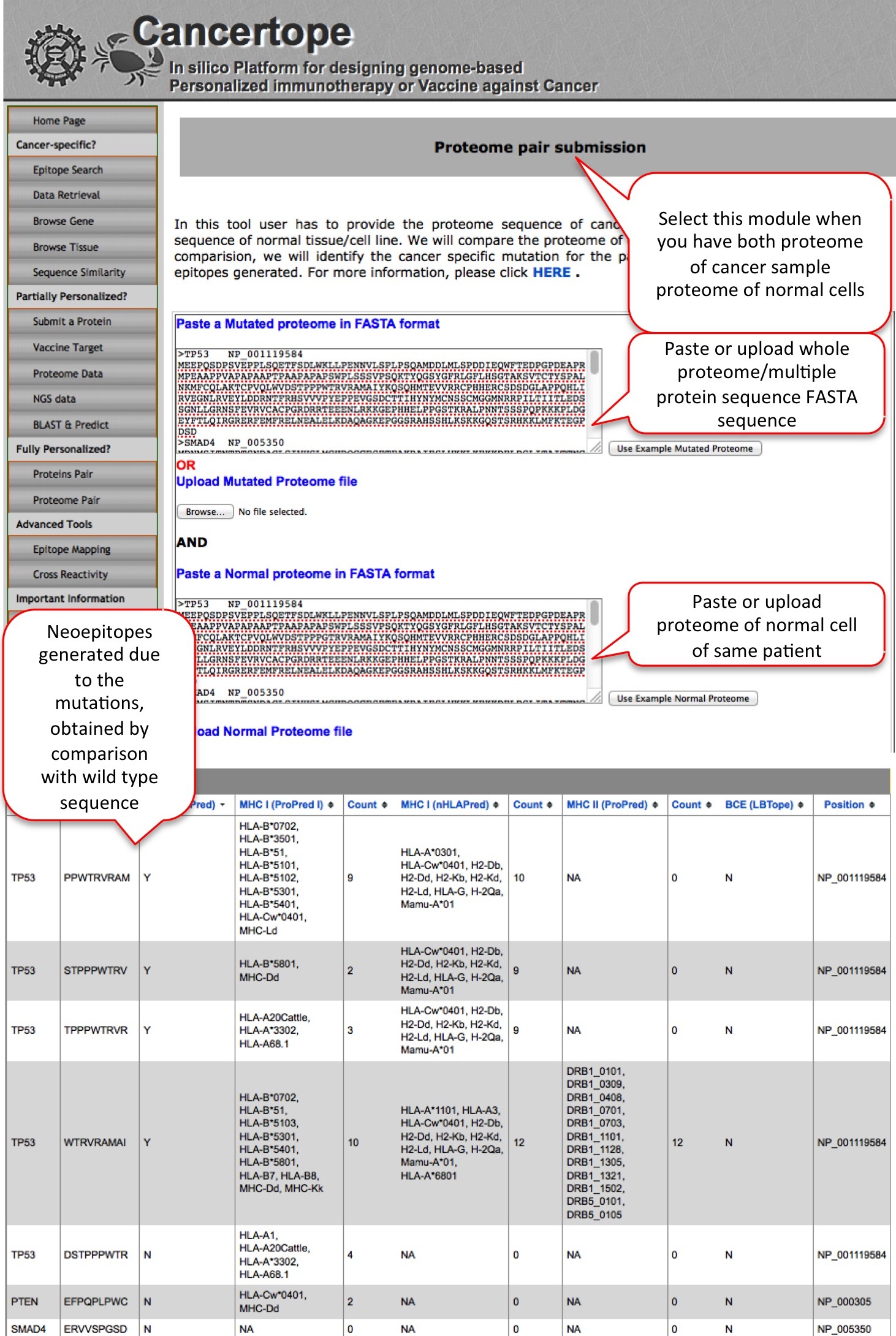 |
Epitope Mapping This module is given for the exploration of experimentaly validated epitope mapping. For example submission of TP53 mutant leads to different experimental immune epitopes from IEDB and MHCBN databases. |
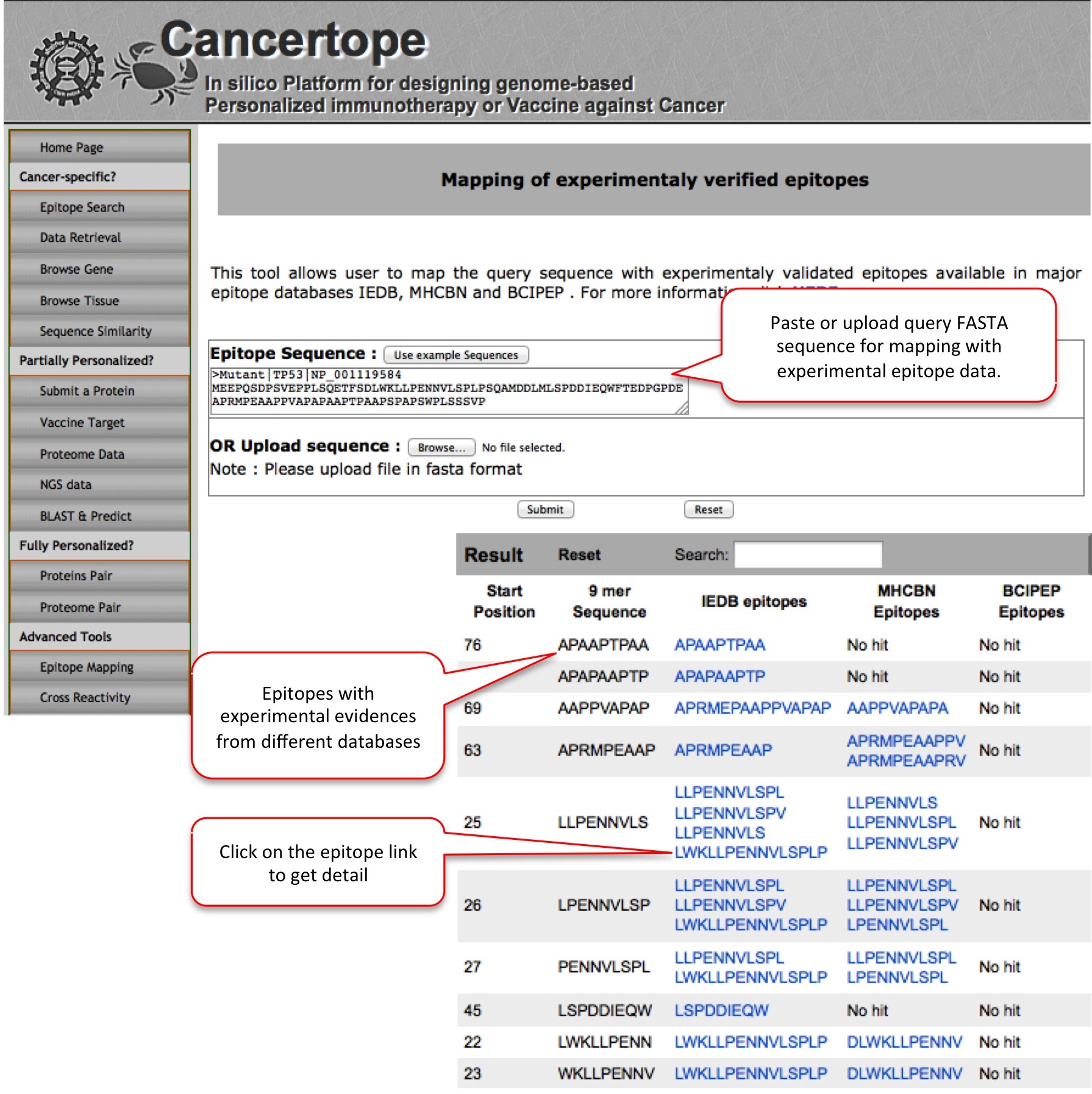 |
Cross Reactivity This module is designed to lessen the possibility of cross reactivity in the candidate in question. FOr example here we have vaccine candidate AAK1 and we want to see the regions in the query, which are not present in the selected dataset such as 1000 genome-based proteomes. We get GLGSGSSGK etc. peptides which are not cross reactive in the 1000 genome-based proteomes. |
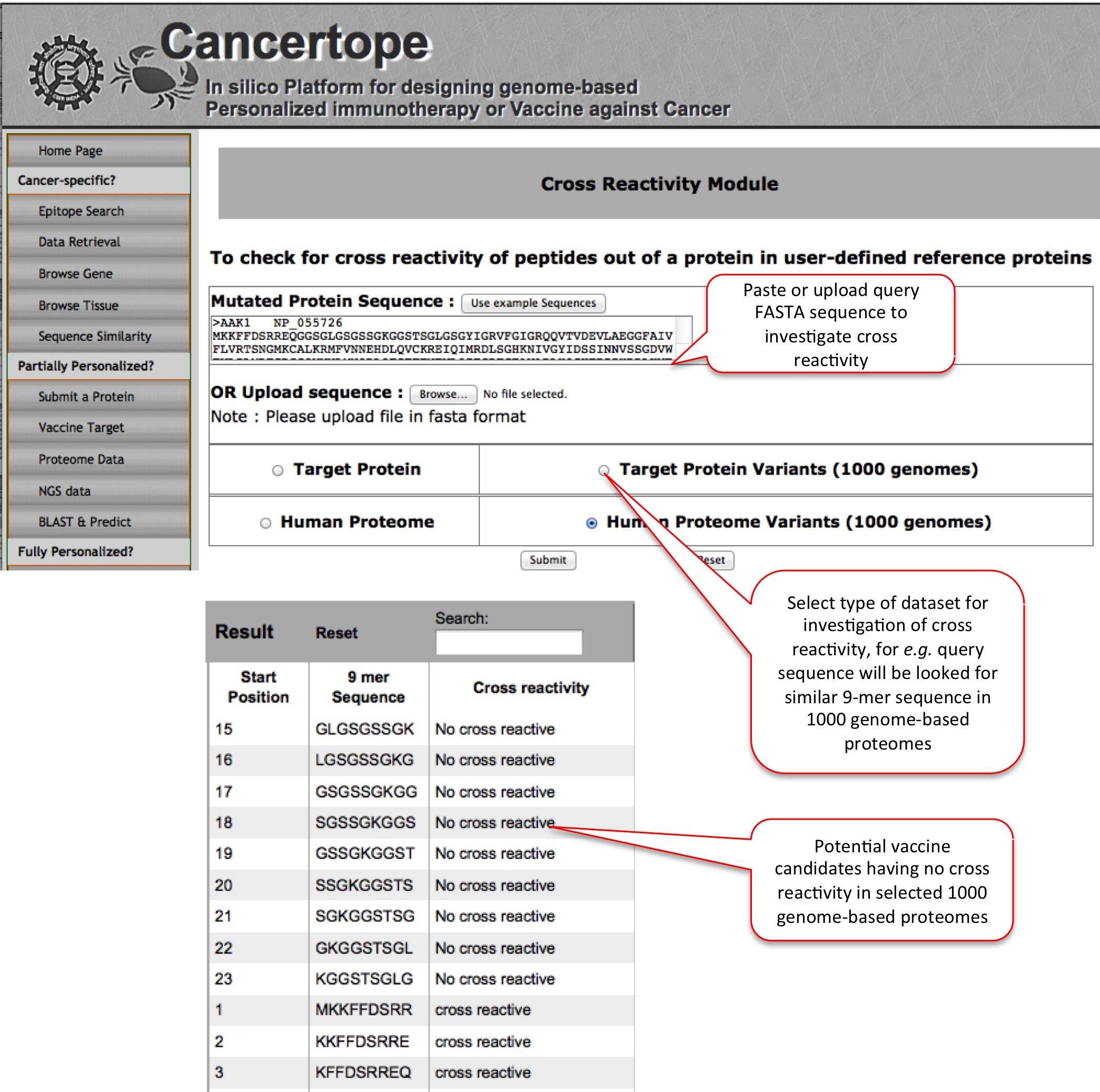 |
|
CANCERTOPE | Raghava's Group | IMTECH | CSIR | CRDD | GPSR Package |