|
IL4pred:This web server is developed for users working in the field of vaccine design. This server allows users to predict and design Interleukin-4 (IL4) inducing peptides. It have following four modules: |
| 1. Peptide Analogs: It generate mutant (analogs) of a query peptide and predict IL4 inducing epitopes in all analogs. This server allows users to identify minimum mutations required to increase its potency. |
Input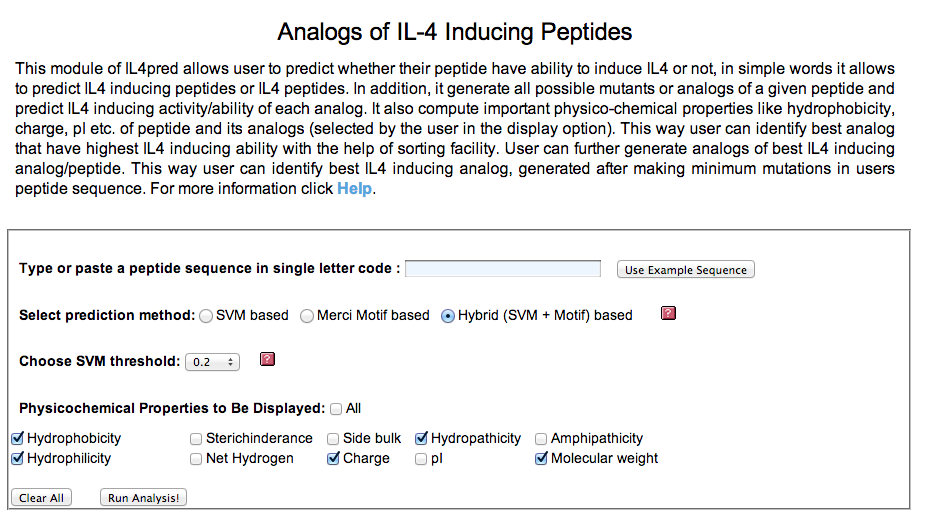
Result 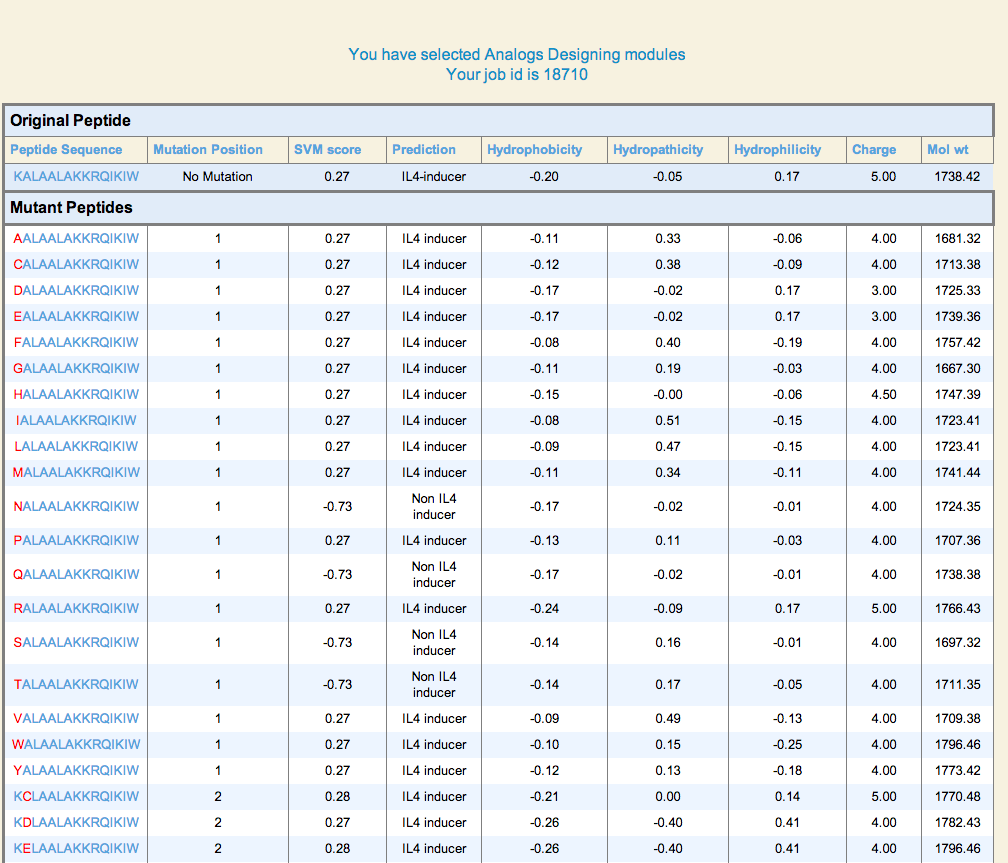
Loop 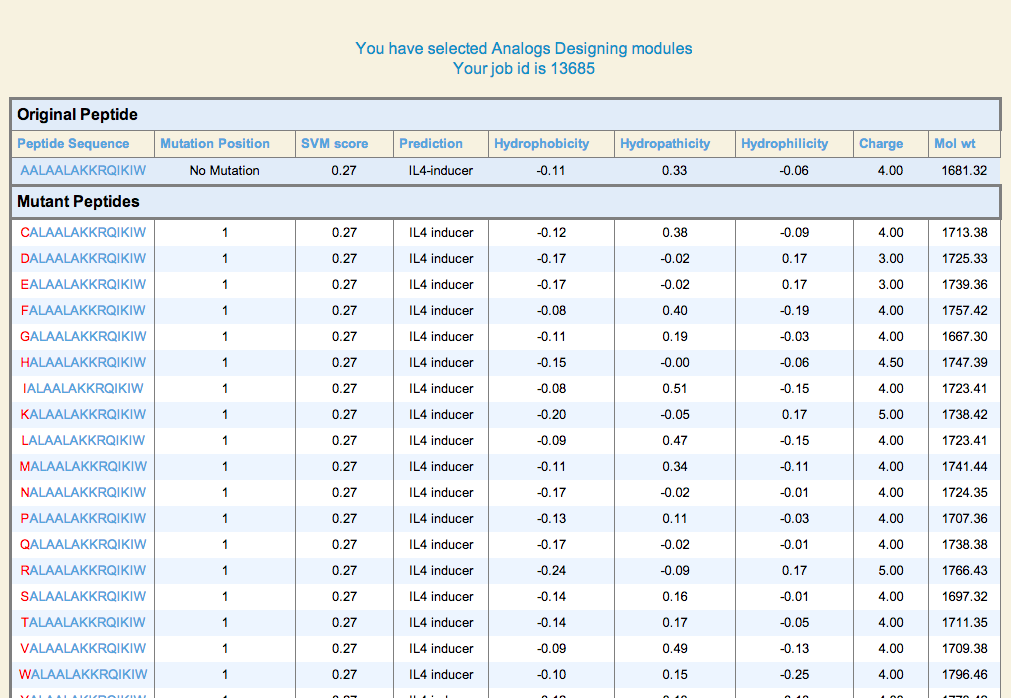
|
| 2. Virtual Screening: This module allows users to predict the IL4 inducing peptide/epitope in a set of peptides or peptide library (virtual screening). User may submit large number of peptides and server will predict and rank them based on their potential to induce IL4. |
input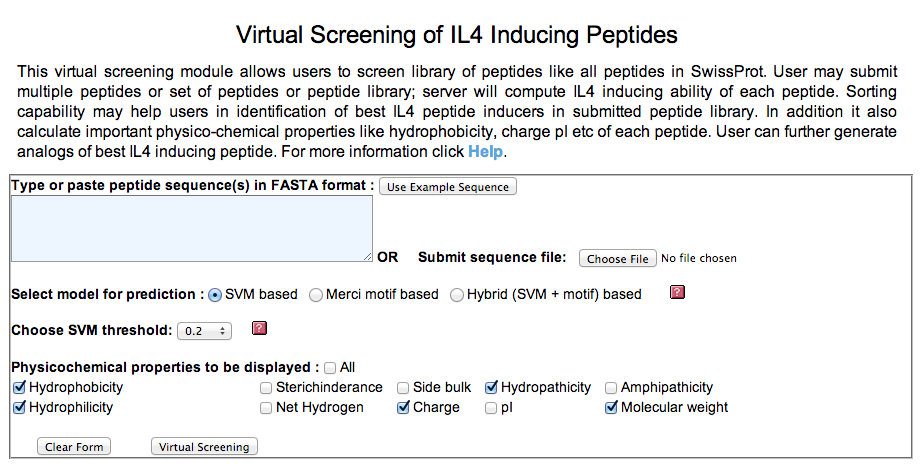
Result 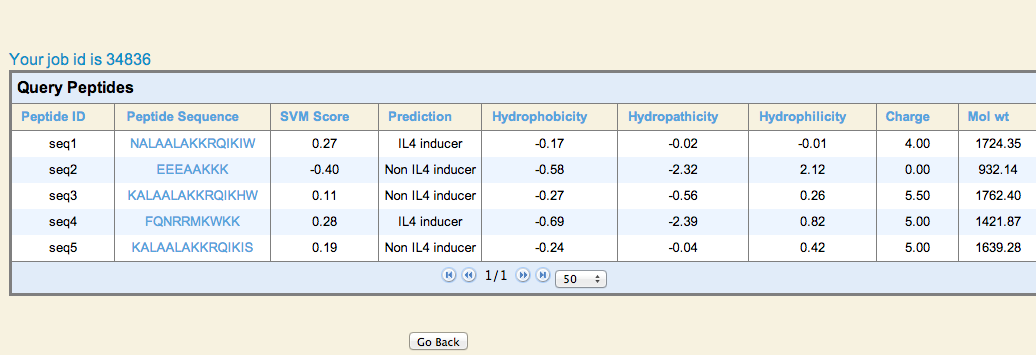
LOOP 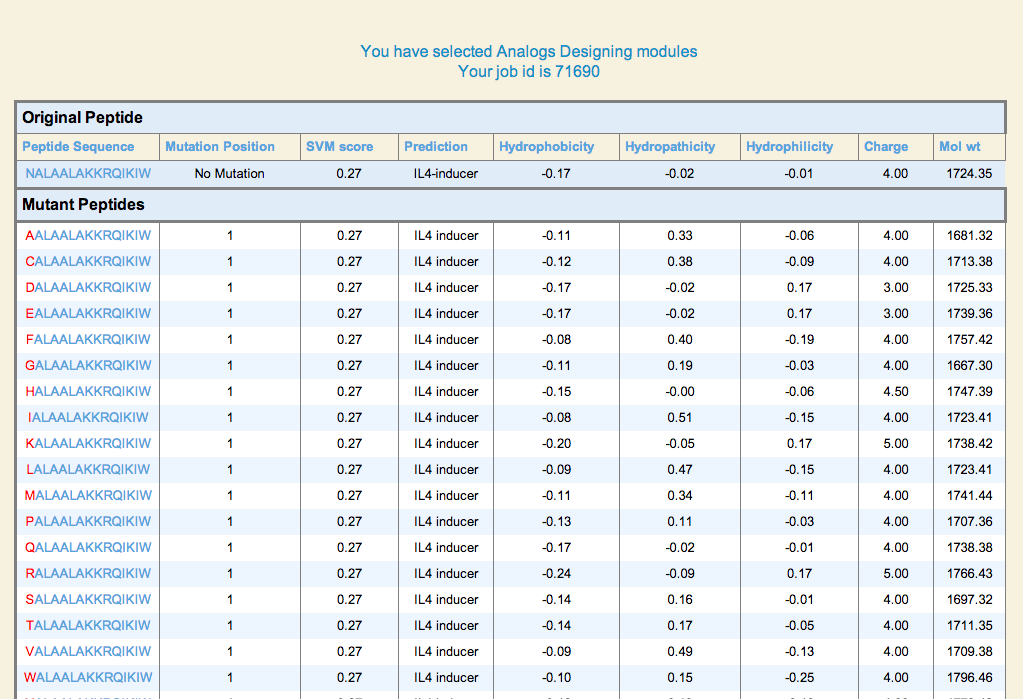
|
| 3. Protein Mapping: This module generates overlapping peptides of the query protein/antigen sequence and predicts IL4 inducting ability of these antigenic regions. It allows users to identify regions in a protein/antigen have potency to induce IL4. |
input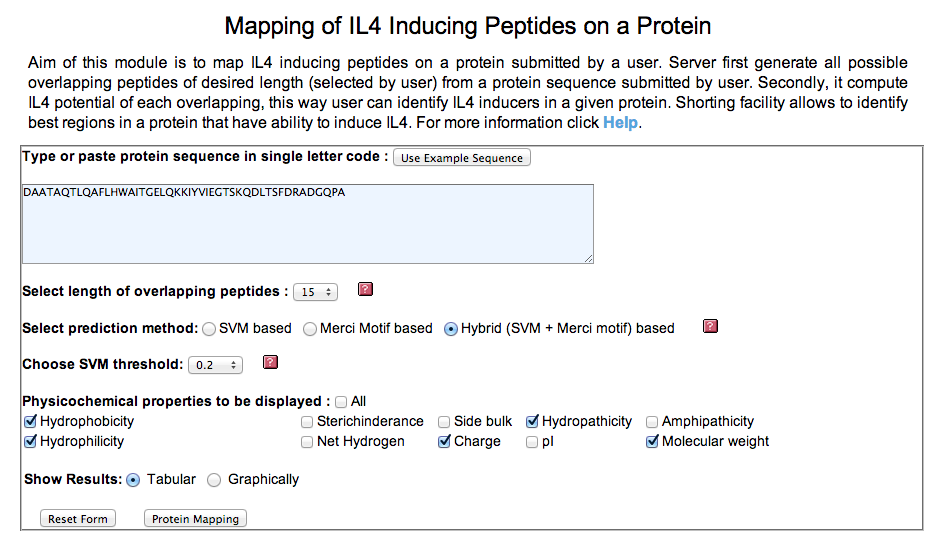
tabular Result 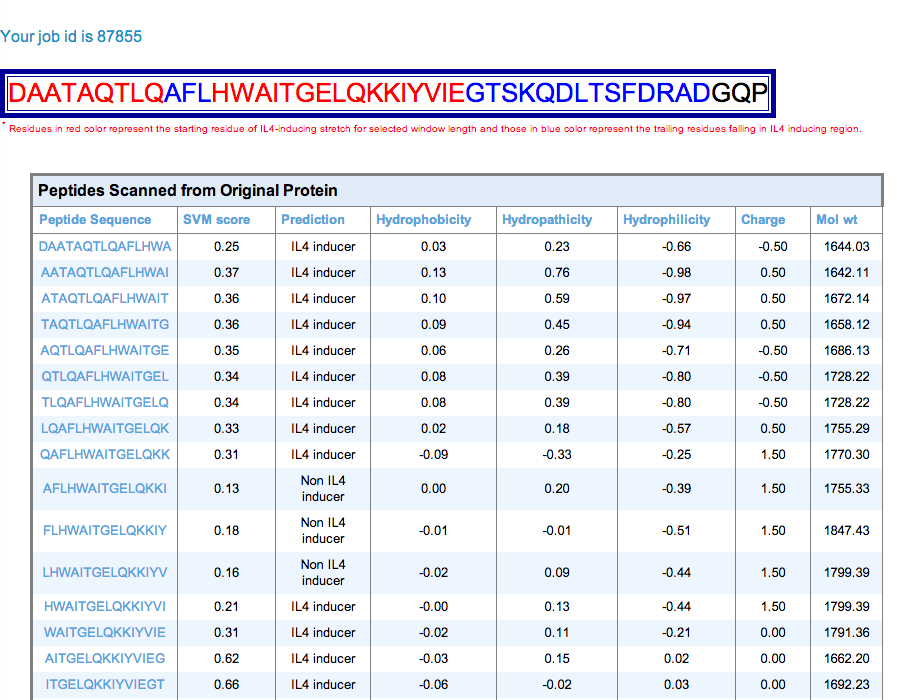
Graphical display 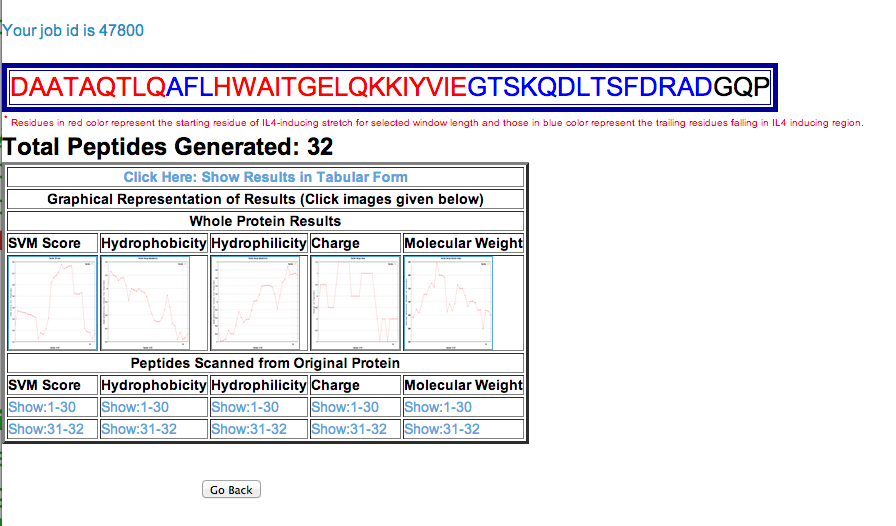
|
| 4. IL4 motifs: Motifs have been shown to play a very important role in discrimating the two datasets. This module will help the user to search the motifs that were found important for IL4 inducing potential. Here, the user can search the motif using MERCI, under three classification of amino acid: a)NONE b)BETTS-RUSSELL c)KOOLMAN-ROHM. |