dbEM
A Database of Epigenetic Modifiers
Cite: Nanda JS, Kumar R, Raghava GP. (2016) dbEM: A database of epigenetic modifiers curated from cancerous and normal genomes. Sci Rep. 6:19340.
|
Epigenetics defines the changes in the gene expression or cellular phenotype (which are heritable) without a change in the nucleotide sequence. The epigenetic mechanisms involve DNA methylation, histone modifications and RNA silencing. Alterations or mis-regulation of gene expression have been associated with a variety of malignant diseases such as cancer. Recent genomes-wide studies on cancer have shown that epigenetic mechanisms play an important role in the development and progression of disease. Epigenetic marks are mainly executed by a variety of proteins that may function as writers (HATs, HMTs), readers (chromodomain binding proteins), and erasers (HDACS, HDMTs/KDMs, phosphatases, deubiquitinases). The activity of these enzymes is further regulated by a variety of chromatin remodeling complexes and their components. In order to cure cancer certain epigenetic enzymes, particularly HDACS, have been targeted and many inhibitors against these have entered drug trials that have met success either alone or in combination with other chemotherapeutic drugs. |
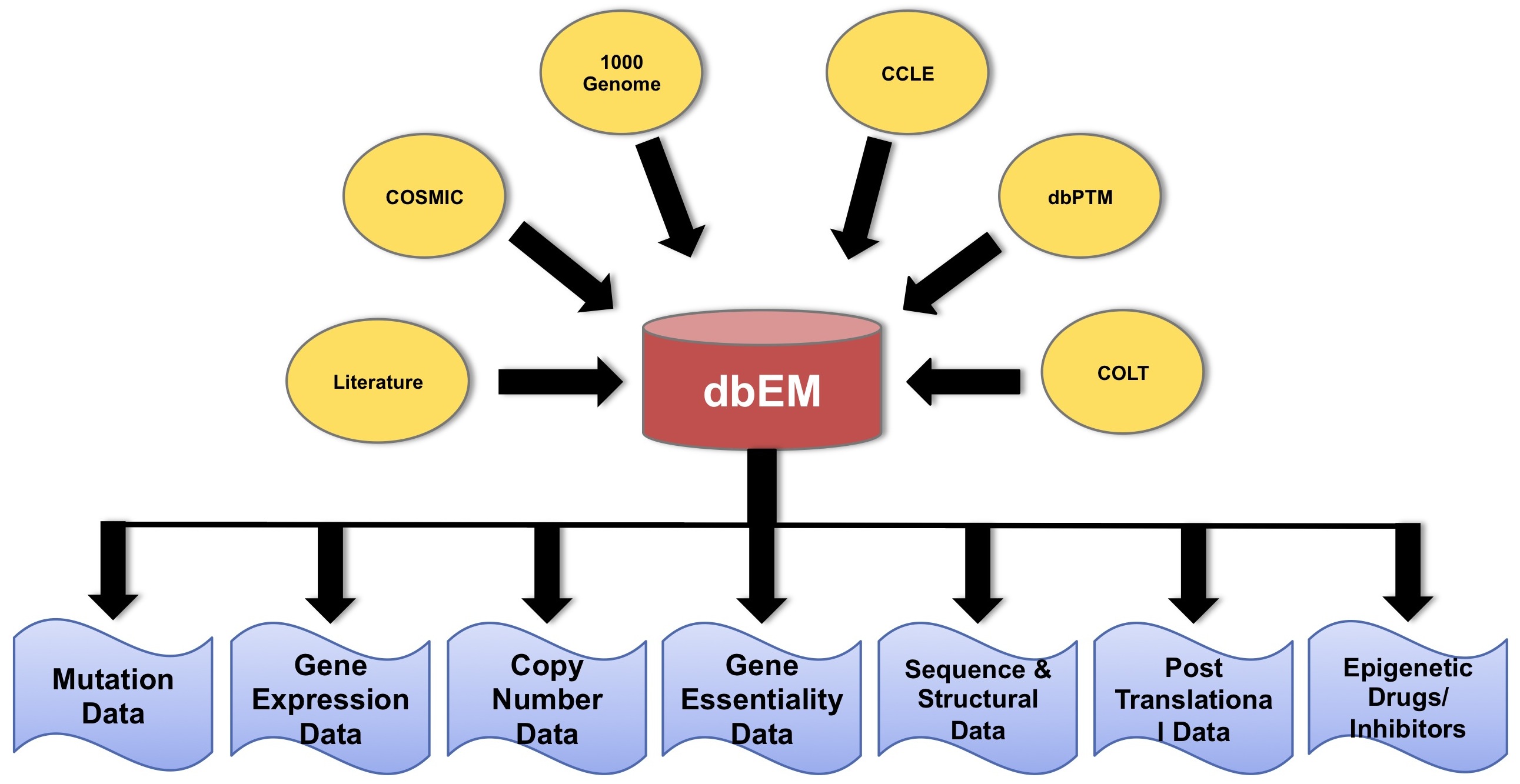 |
In the light of above findings we have tried to made a database "EPICANDB" for highlighting the role of various epigenetic modifiers in cancer. This database contains 168 proteins that have an epigenetic function. We have extracted the mutations, expression level and copy number variations of their genes across thousand of cancer cell lines and tumor samples. Apart from this database provides some additional features including the gene essentiality data, sequence alignments of these proteins with mutated sequences and their homologous proteins. Structures for those proteins that do not have PDB entries were also predicted. It also gives information regarding PFAM and Superfamily domains present in these proteins. This database even includes information about some epigenetic modifiers used as drugs to treat cancer. This database contains crucial information related to various epigenetic proteins and their relevance in cancer. |