Help page of PCMdb |
| Go to Frequently Asked Questions to know about the purpose and rationale of PCMdb. |
| Abbreviations used in PCMdb for Pancreatic Cancer Types |
|---|
| Adeno | Adenocarcinoma | Muc_neo | Mucinous Neoplasms | Endo | Endocrine |
| Oth | Others | S_not | Subtype Not Mentioned |
SEARCH | |||||
This is a very simple and easy search option. User can search any query in any of the fields (e.g. Gene ADRB3).Results will be displayed according to the selected fields to be dispalyed. e.g. Gene Name, PMID, Cell Line/Tissue, Methylation status etc. | |||||
This search has multiple options which give very refined result for the user. Here user has been provided with different search options like Field name; Match option (equal =, less <, greater > ). Value column is provided to add input from user e.g. Gene Name, Methylation status, Cancer, etc. and then user can apply conditions(AND & OR). 1. If user is giving GENE NAME in the field Gene and wants to match it with ABCB1, the user will opt for (LIKE), result page will display all the enteries containing ABCB1 in the field Gene. 2. If condition AND is applied then another search field will open and user can put Methylation Status like High Level Methylation in Methylation Category field, then results with Gene Name ABCB1 reporting High Level Methylation will be displayed. 3. If condition OR is applied with the above input, then the user will get all enteries containing either Gene Name ABCB1 or will have methylation category High Level Methylation . 4. Match option LIKE, if user is opting for LIKE option then value must be put in between two % symbols e.g. %GGTC%. Here user will get result searched for GGTC in the given field option, if the user is putting % in front then result will be displayed where the word GGTC is in the beginning and if it is put after word like GGTC%, then user will get result where GGTC word is in the end of the field which is choosen. *Note: While submitting, the advanced search for various conditons, the last search box in the conditions must be blank. By pressing search, desired result will be displayed. | |||||
BROWSE | |||||
This interface allows user to browse the Cell Line field in the PCMdb database.
| |||||
In this page the user can browse by gene name by clicking on the first alphabet of the gene name.
| |||||
User can browse and obtain PCMdb entries for a particular category of methods used for reporting the methylation status of the gene in thesource research article.
| |||||
User can access entries of PCMdb based on methylation status namely "High","Low","Intermediate","Altered" or "Unknown".
| |||||
User can browse and obtain PCMdb entries for the genes located on a particular chromosome number.
| |||||
SUMMARY | |||||
This page provides comprehensive information about genes with sortable columns of number of entries, cell line/tissue samples, methylation status, and number of studies.
| |||||
This page provides the major categories of gene functions to which the genes in PCMdb belong to.
| |||||
This page gives a list of genes that have been reported to be potential biomarkers in cancers other than pancreatic cancer as free nucleic acid fragments originated from these genes and circulate in blood and the methylation status of these genes can be found out from these circulating free nucleic acid fragments.
| |||||
This page provides chromosome wise summary of data entries in PCMdb.
| |||||
DRUG RESISTANCE | |||||
At this page, we are providing methylation status of genes. These genes methylation have been reported to be involved in drug resistance.
| |||||
At this page, we are providing summary of methylation status of pancreatic cancer cell lines presnt in CancerDR.
| |||||
This page provides the drug sensitivity status of different pancreatic cancer (PCa) cell lines (obtained for CancerDR database), along with the methylation status of their drug targets in our database.
| |||||
TOOLS | |||||
User can run a BLAST search for sequences. The server provides options to choose different type of BLAST types and expectation value.
| |||||
Smith-Waterman Search page assists the user to run a Local alignment of sequences using Smith-Waterman algorithm.
| |||||
This module will be used for the alignment of short reads, produced after the Next Generation Sequencing (NGS) of genome, Transcriptome or Exome. Both types of sequencing reads i.e Paired end and single end sequencing reads can be submitted to this module. Sequencing reads will be aligned to the methylation target genes and visualized for any variation. Alignment visualization will clearly indicate the mutations in the sequenced genes. User has to provide the files (.fastq format) generated after the sequencing (Please see the example files). This module use BWA to generate alignment files (i.e. aln.sam file) and Tablet for visualization of the alignment files. It is necessary to provide forward and reversed read files both at a time if a user wants to use paired end data for the alignment. A single file is enough in case of single end sequencing.
| |||||
|
User can submit Whole genome assembly of Next Generation sequencing (NGS) data generated contigs to this module. This module predicts the genes from contigs and align to the methylation target genes.
| |||||
This page contains a form in which the user can select cell lines for comparison of the methylation status of genes in different pancreatic cancer cell lines.
| |||||
GUIDE/HELP | |||||
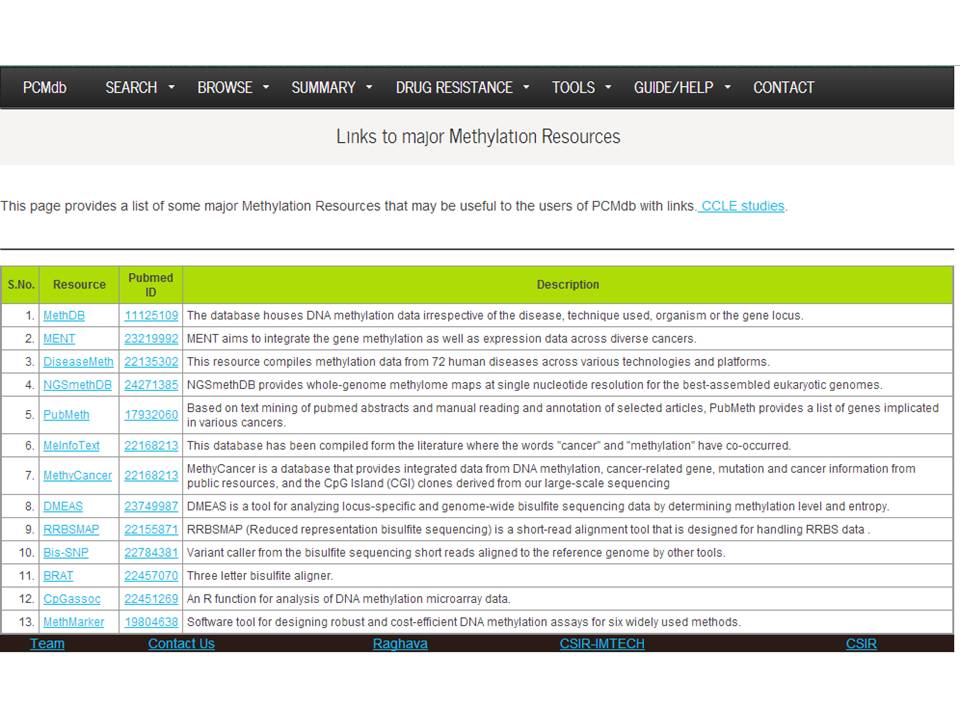
This page provides a list of some major Methylation Resources that may be useful to the users of PCMdb with links. | |||||
 | |||||
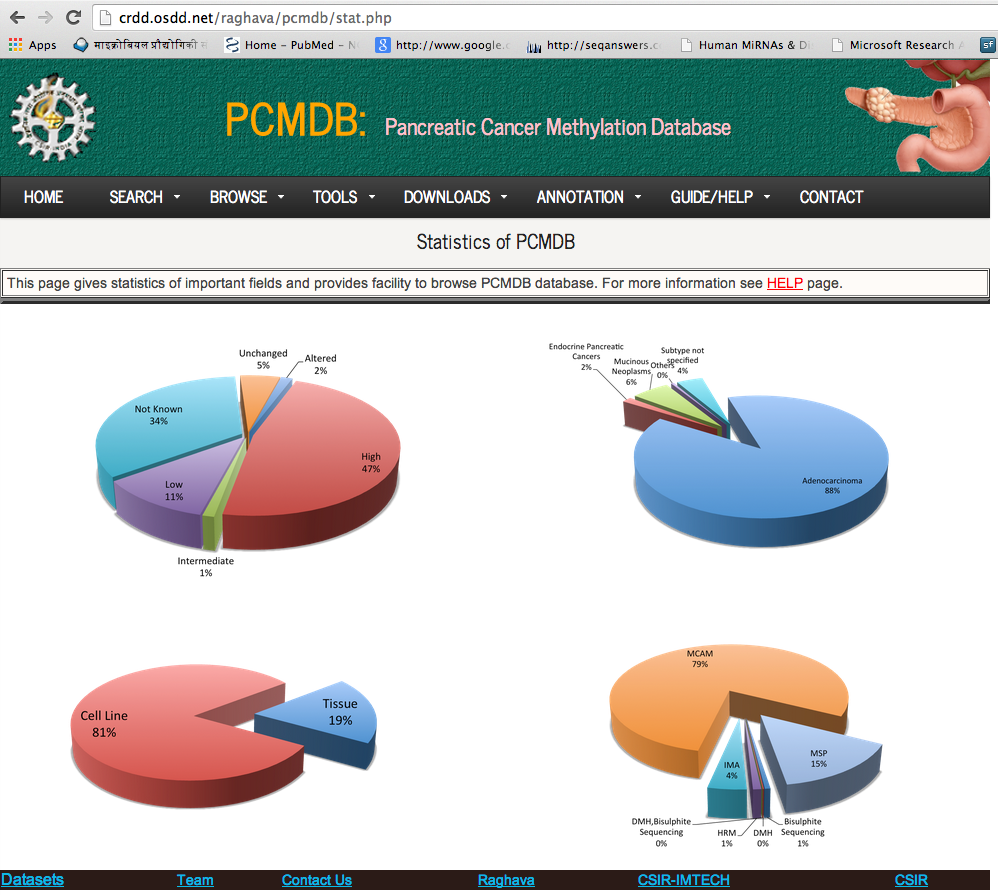
This page gives statistics of important fields and provides facility to browse PCMdb database.
| |||||
 | |||||
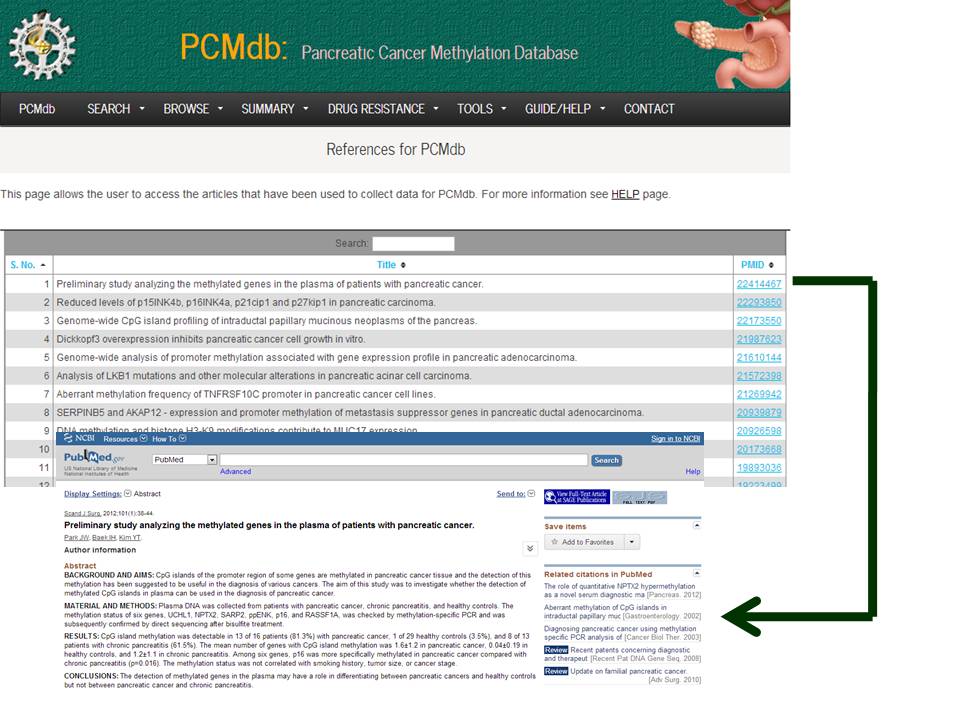
This page allows the user to access the articles that have been used to collect data for PCMdb.
| |||||
 | |||||
Q1. What is PCMdb? Ans. PCMdb is a compilation of methylation status of the human genes in pancreatic cancer collected from both single gene studies as well as high throughput genome wide studies. Q2. What is DNA methylation and how is it related to cancer? Ans . A large number of genes in vertebrate genomes have short CpG rich regions called CpG islands (CGIs) and the rest of the genome is deficient in CpGs in general. Much of the work done in DNA methylation has focussed mainly on CGIs at the transcription start sites or promoters of the genes. Methylation of cytosine at carbon 5 position is an epigenetic phenomenon commonly found in many eukaryotes. When methylation occurs at CGIs of the promoter regions of genes it is usually thought to be a repressive condition with respect to the gene expression. Though exact mechanisms of this effect still remain to be elucidated, it is known that dedicated DNA methyltransferases with well conserved catalytic domains are present to cause CpG methylation. In cancers, epigenetic silencing of tumor suppressor genes by virtue of excessive or hypermethylation at promoter region plays a definite role in pathogenesis and progression. Since epigenetic processes like DNA methylation are reversible, methylation inhibitors likeazacitidine and decitabine are being increasingly studied for their anticancer application. Q3. Why PCMdb Database is created? Ans. Disruption of methylation has been recognized as one of the key players in cancer development and progression. Many crucial genes like tumor supressors and oncogenes have an altered methylation state in cancer.These can serve as potential methylation based genomic markers and aid in early detection and diagnosis of fatal diseases like pancreatic cancer.Pancreatic cancer is one of the leading causes of cancer deaths worldwide.Therefor,we have catalogued experimentally validated genes and their methylation states in all types of pancreatic cancer. Q4. What is unique about PCMdb? Ans. One of the key features of this database is the analysis that the user can make between the drug sensitivity/resistance data for different cell lines/genes with the methylation state of of each gene in pancreatic cancer.Drug resistance is one of the main reasons behind limited anti-cancer therapies.So we have extracted the drug resistance data from the CancerDR database for different cell lines/genes and correlated it with the methylation data for each gene in pancreatic cancer.The user can also compare the data on different pancreatic cancer cell lines. Q5. Does this database represent all the methylation data available for pancreatic cancer available in literature? Ans. This database is the result of first round curation and the literature is being continuously searched for more studies with the objective of providing an exhaustive repository. Q6. Why search PCMdb? Ans. PCMdb provides all the experimenatlly validated information about genes such as their methylation status,methylation value,experimental details like type of sample used(cell line/tissue),technique,control sample used and cancer type.Along with this,the user can also access the data on expression of important genes, mutation status,copy number variations and drug resistance/senstivity data, hence correlating it with the methylation status of genes in pancreatic cancer. Q7. How do I process a text search with PCMdb database? Ans. User can search a gene by gene name,methylation status,mathylation technique,cell line,cancer type,PMID,etc. Q8. Is this database useful if users have their own query sequence? Ans. Yes, user can use tools like BLAST, SMITH-WATERMAN, short read mapping, due to which sequence analysis is possible in-house. Q9. To whom can I report a discrepancy? Ans. Please refer to the "Contact Us" page. | |||||
| Team | Contact Us | Raghava | CSIR-IMTECH | CSIR |
