 Introduction
Introduction
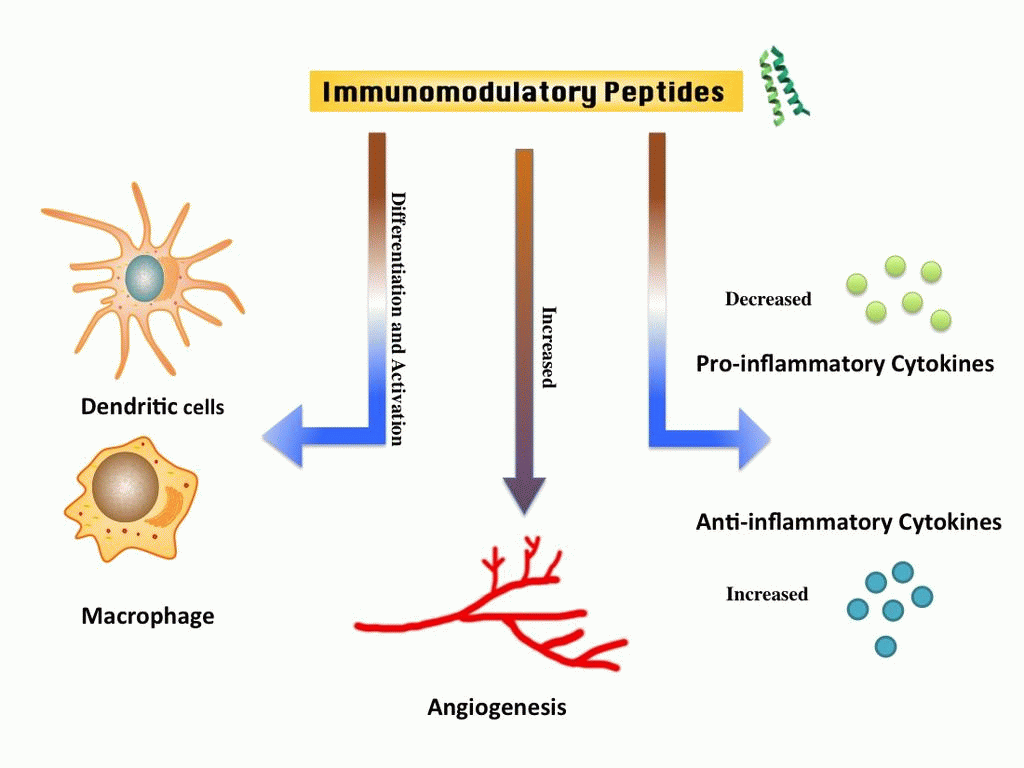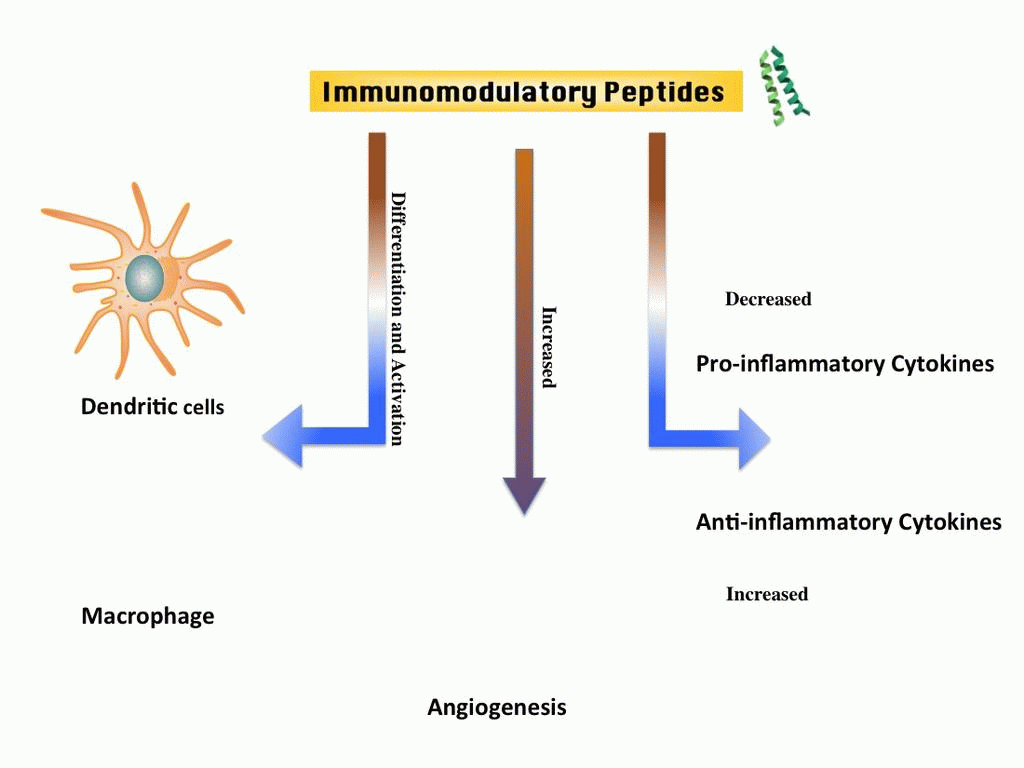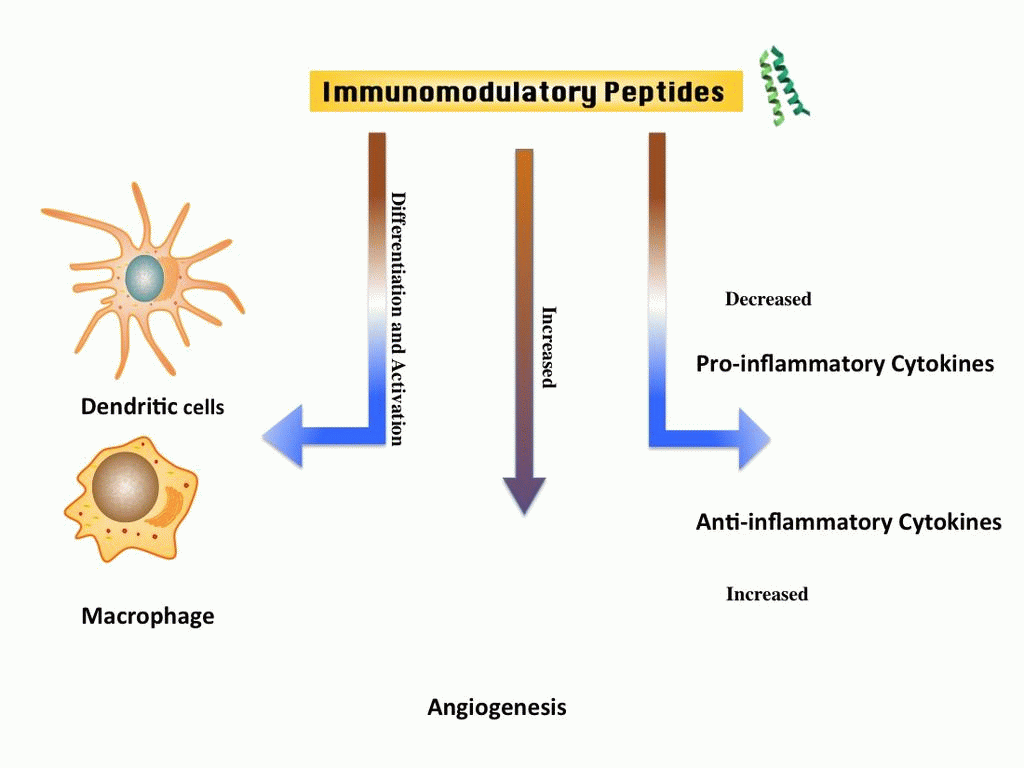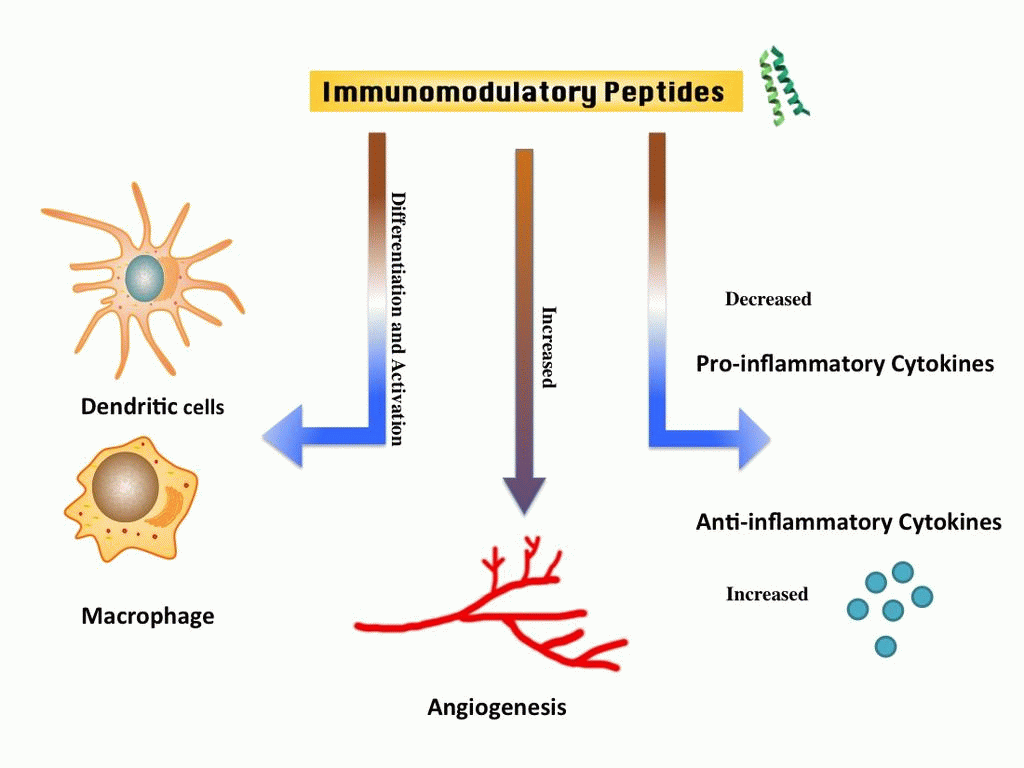
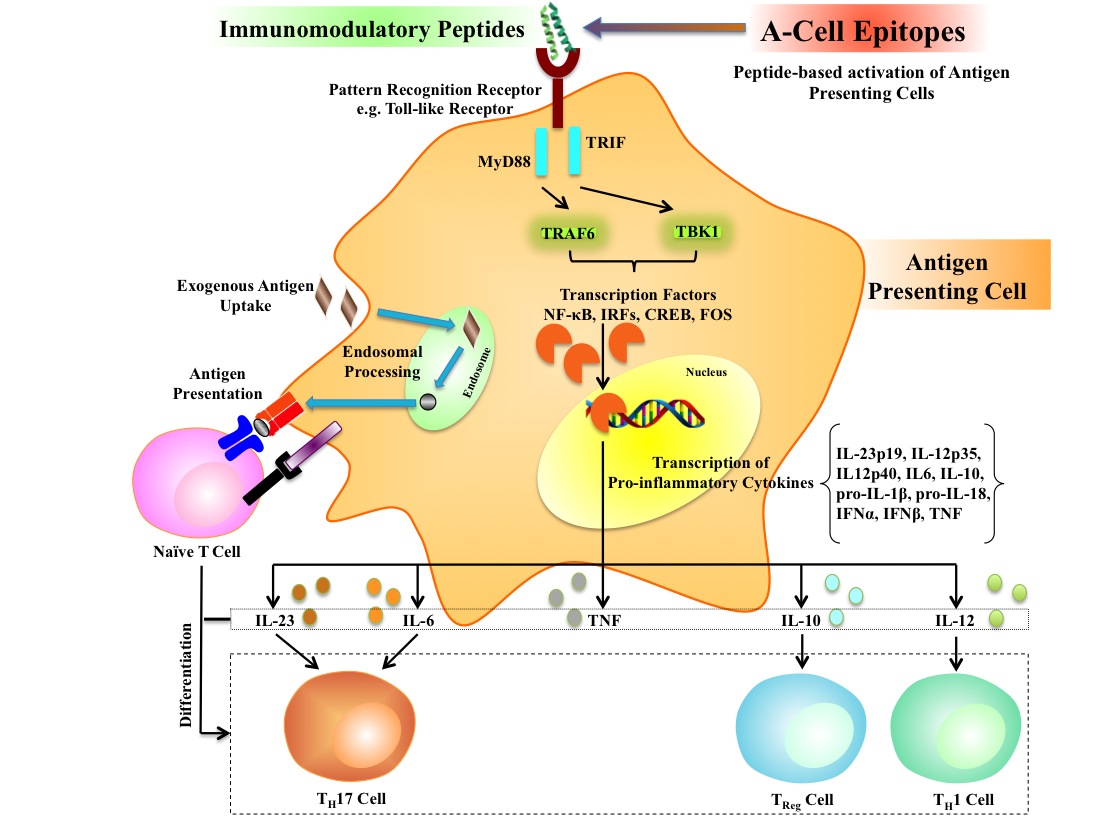
In past, numerous methods have been developed to predict B-cell and T-cell epitopes for designing peptide based subunit vaccine. Aim of these epitope prediction methods is to identify peptides (antigenic regions in proteins) that can acticate or boostadaptive arm of immune system. In contrast no in silico method has been developed to predict peptide that can activate innate immunity; no method for designing peptide based vaccine adjuvants. It has been shown in previous studies that toll-like receptors recognize peptides and can activate antigen presenting cells. It means peptides have potential to act as vaccine adjuvants as they have immunomodulatory potential to activate innate immune system. In this study we called these peptides as A-cell epitopes as they have ability to activate antigen presenting cells (A-cells).
Experimentally validated immunomomodulatory (A-cell epitopes) and non-immunomodulatory peptides were collected and compiled from various resources. We used these experimentally validated peptides for developing SVM-based models for predicting A-cell epitopes using different sequence-based features of peptides. These models were implemented in a web-based platform VaxinPAD for designing peptide-based vaccine.
 Following are major faetures have been incorporated in VaxinPAD to faciliate community in designing peptide based adjuvants.
Following are major faetures have been incorporated in VaxinPAD to faciliate community in designing peptide based adjuvants.
Prediction of A-cell Epitopes: This module allow user to predict immunomodulatory peptides or A-cell epitopes (Antigen presenting Cell epitopes). It also allow users to perform virtual screening of peptide librarries to identify A-cell epitopes.
Analog of Peptides: This module generate all possible analog/mutants of a given peptide (all possible single mutations) and predicting their respective immunomodulatory potential. This module is developed for designing better vacine adjuvants.
Protein Ajuvatns : This option allow users to predict potential protein-based ajuvants. It identify poteintial A-cell epitopes in a query proteins sequence.
DataSet : The peptide sequences that have been experimentally found to be immunomodulatory in literature have been collected and tabulated here in the form of dataset. Each sequence has been linked to a datacard that provides additional information.