Method
Dataset
PlifePred is developed using 261 peptides whose half-life had been experimentally validated in mammalian blood, collected from PEPlife database. Of these 261 peptides, 98 peptides had chemical modifications or non-natural amino-acids like methylation, aminoisobutyric acid, norleucine, etc. So, we made 2 datasets-
(1) 261 peptides having both natural and chemically modified sequences
(2) 163 peptides having only natural amino acids and no modifications
Features which were used for models which are integrated in the PlifePred webserver-
[1] Amino Acid Composition : It is the percentage content of each amino acid in a peptide/protein and is represented by a vector of 20 values for 20 natural amino acids calculated by the formula
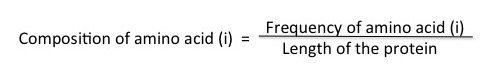
Where "i" can be any natural amino acid.
[2] Chemical Descriptors : Around 15,400 types of descriptors, including 2D, 3D and fingerprints were calculated using the PaDEL software and only the minimum number of important descriptors were used for model building.
Software Packages Used
SVM-Light: Support Vector Machine was implemented using freely downloadable software package SVM-Light written by Joachims (Joachims et al. 1999). This package is powerful as well as user-friendly where we can adjust the parameters and kernel functions like Linear, Polynomial, RBF and Sigmoid.
Weka: Weka a Java based software was used to select features which showed high correlation to half-life and implement SMOreg, Gaussian Processes, IBk etc.
Evaluation and Cross-Validation of Regression Models: The evaluation of accuracy of prediction method is necessary to estimate the performance of a method.The Leave One Out Cross-Validation (LOOCV) procedure was employed to estimate the performance of the prediction methods. The LOOCV procedure involves removing of one peptide from the training data; training is done on the basis of remaining data and then testing was done on this removed peptide. It is the most accurate way to estimate the performance of method when the training data is small.
In order to evaluate performance of our models, we used Pearson’s correlation coefficient (R).
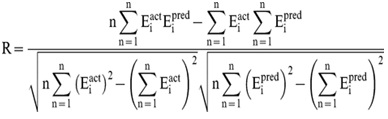
Where n is the size of test set, Eipred and Eiact is the predicted and actual half-life values respectively.
