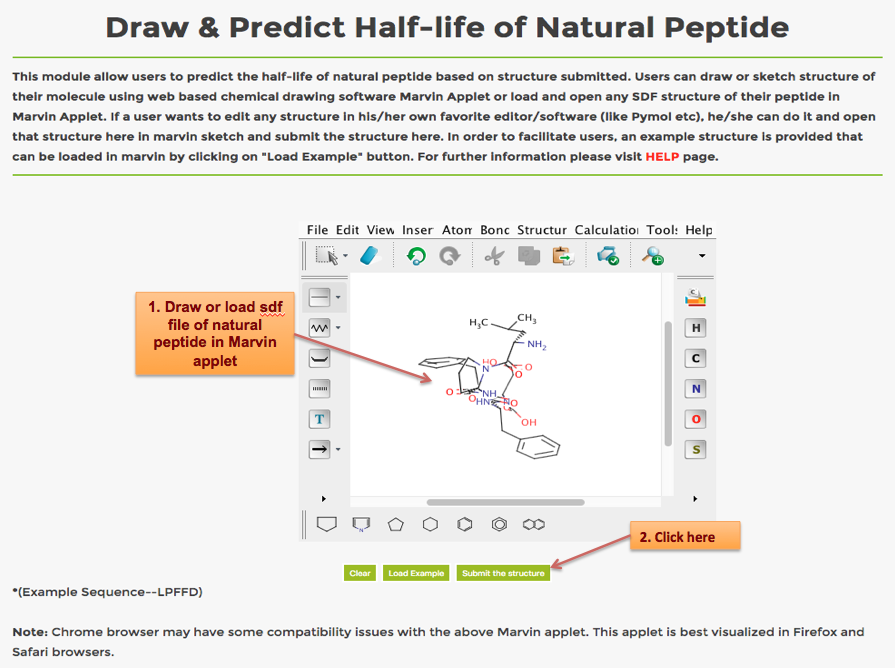
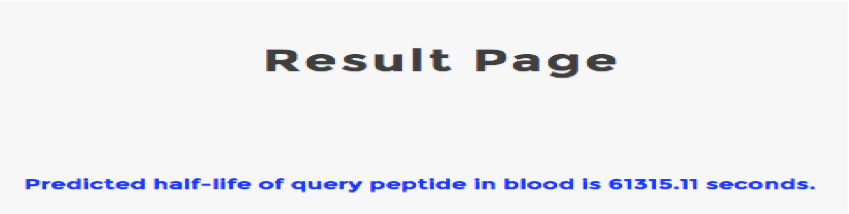
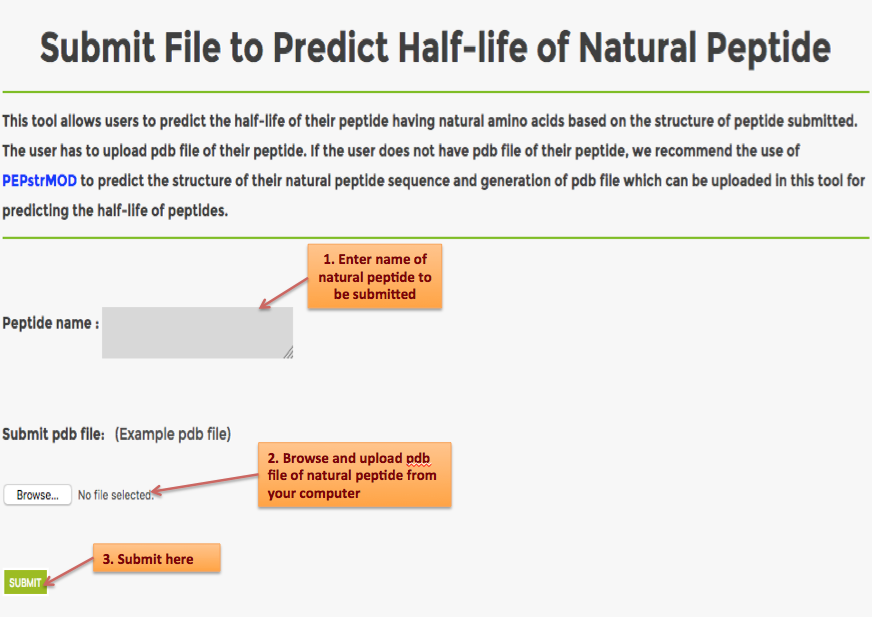
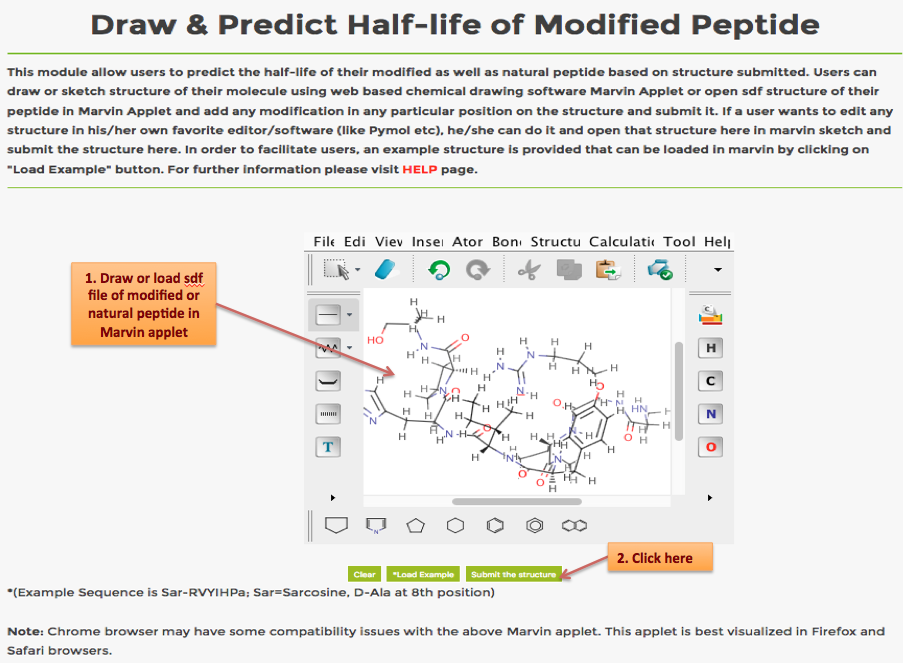
Help
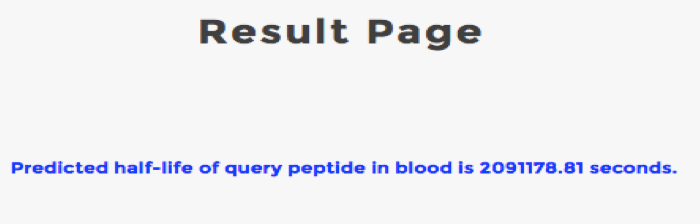
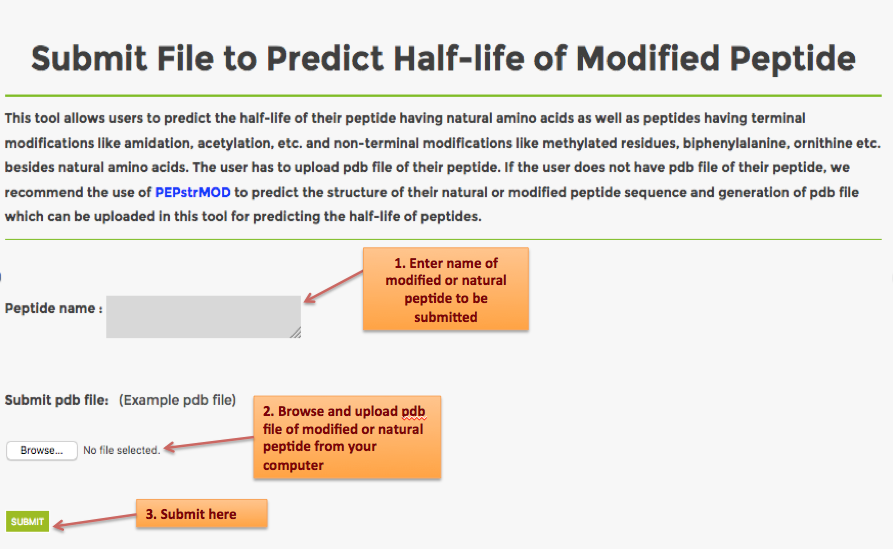
PlifePred is prediction server for predicting the half-life of peptides in blood. Users can use PlifePred to increase or decrease the half-life of their peptide by generating mutants The server facilitates designing the half-life of a peptide by generating its mutants and predicting their half-life and physiochemical properties.The user can screen and select the peptide with suitable properties desired.
| NATURAL | MODIFIED | ||||
|---|---|---|---|---|---|
|
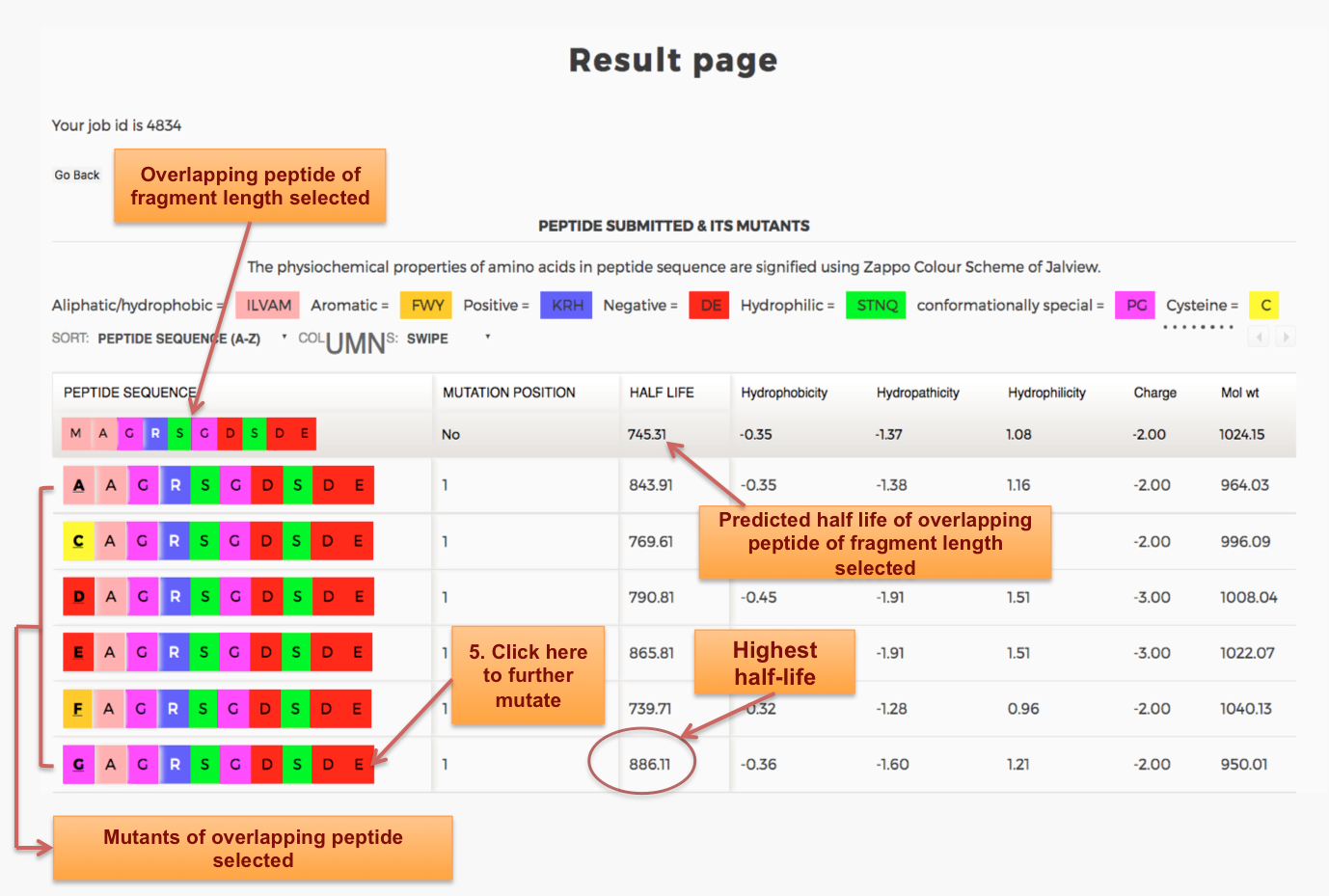
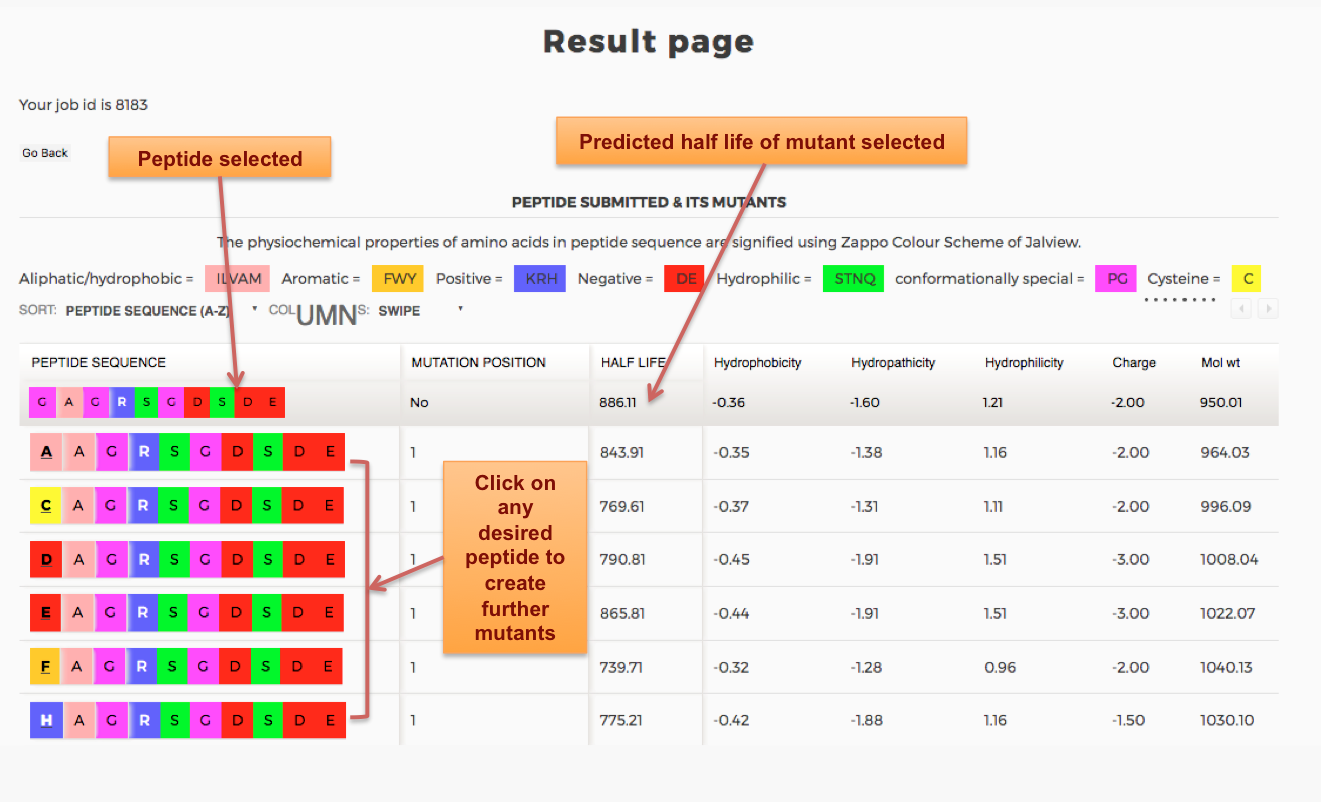
Designing of Half-life of Peptide using Analog Generation Module of PlifePred
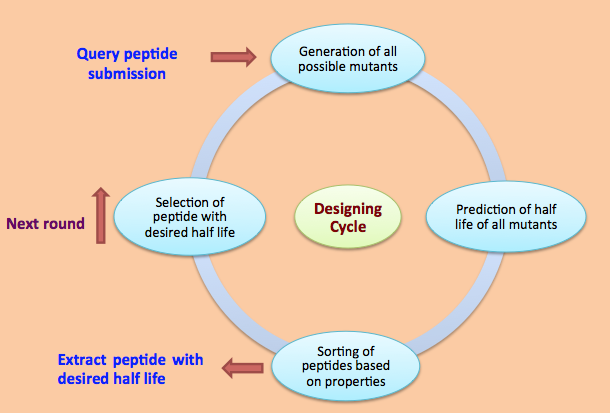
Example demonstrating use of Analog Generation for Designing Half-life of Peptides
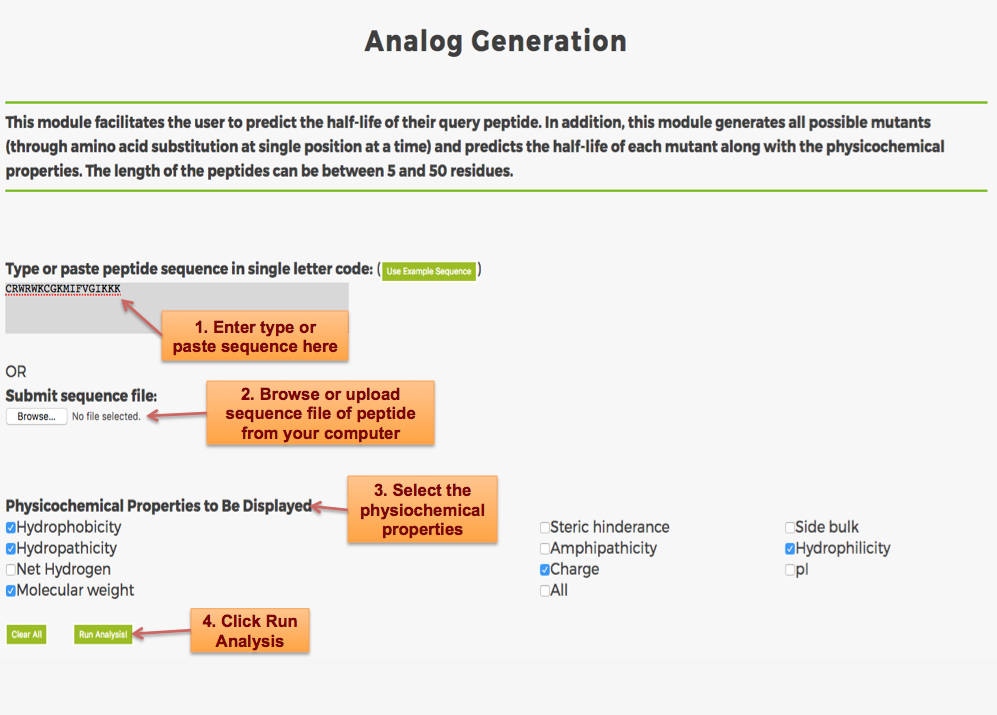
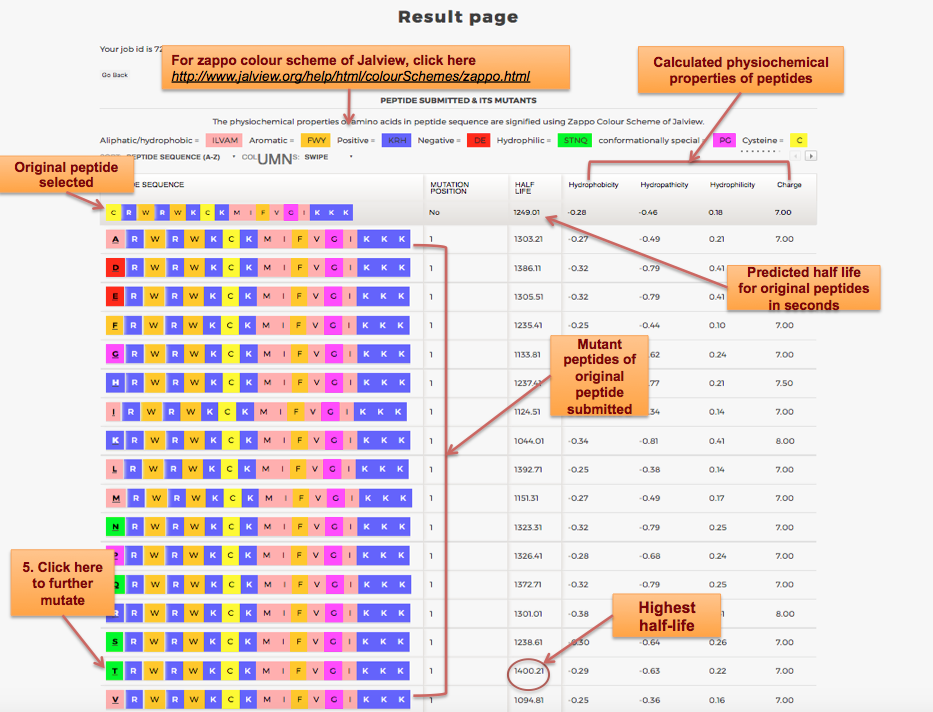
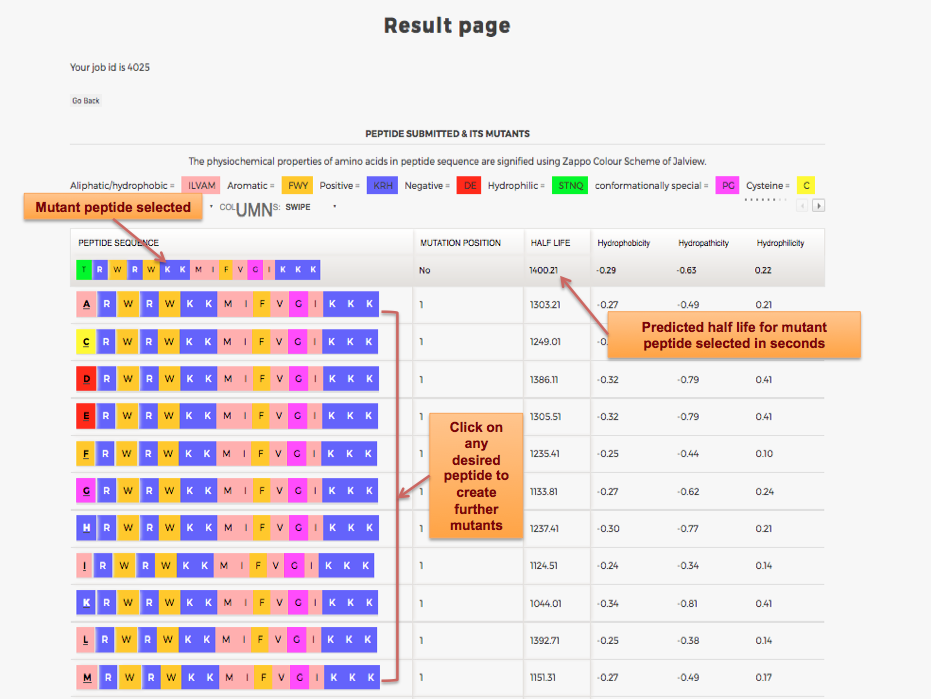
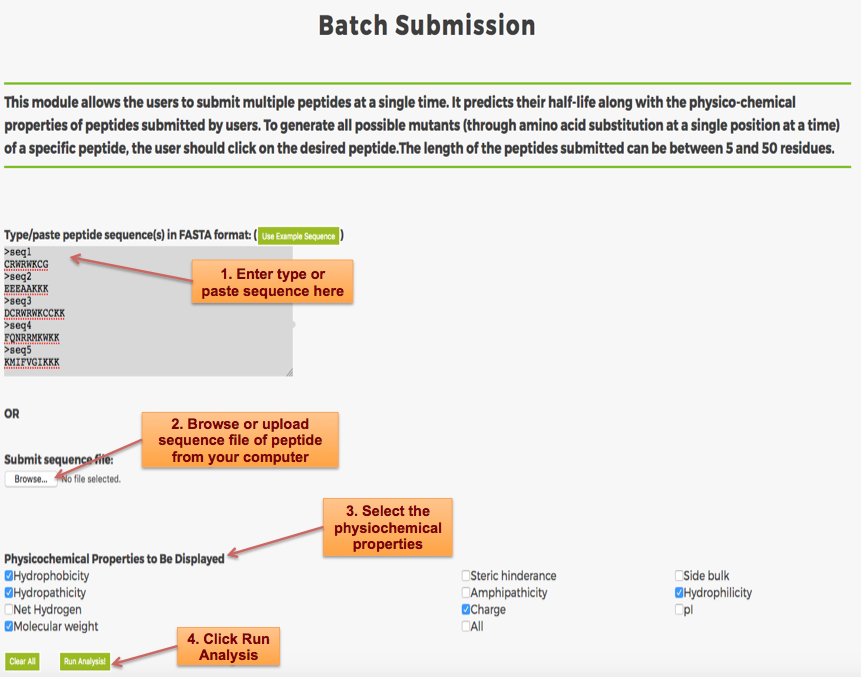
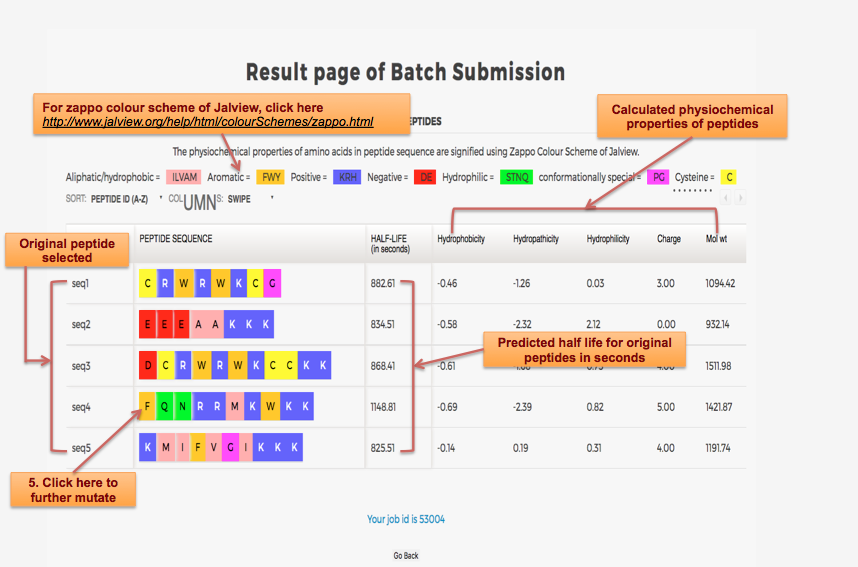
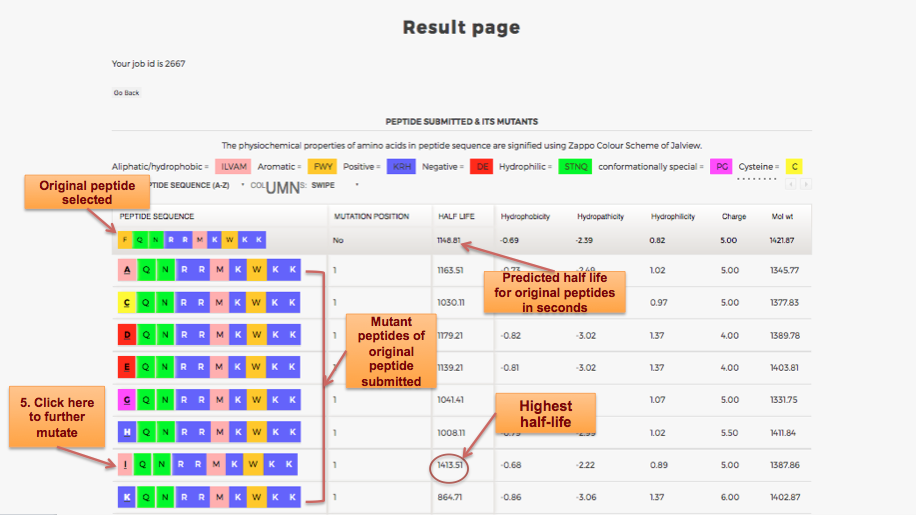
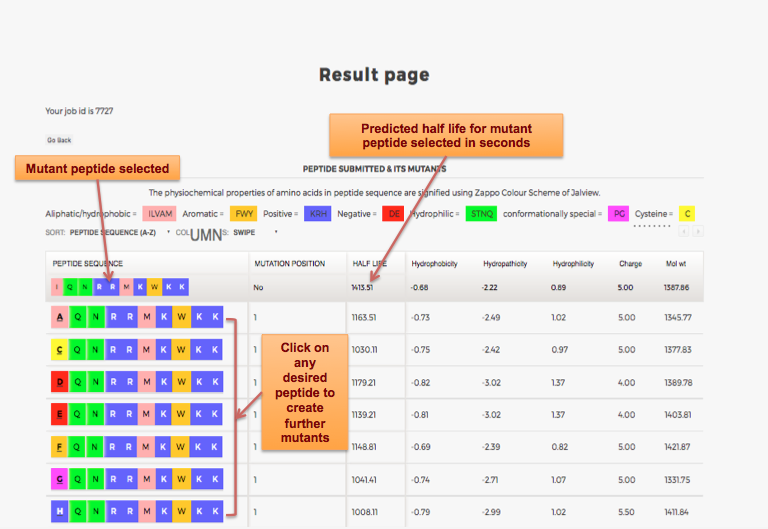
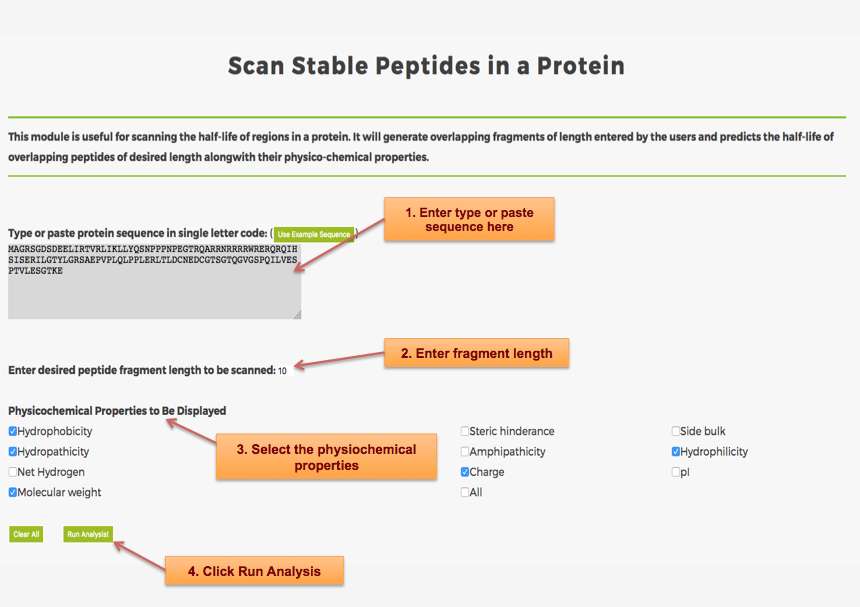
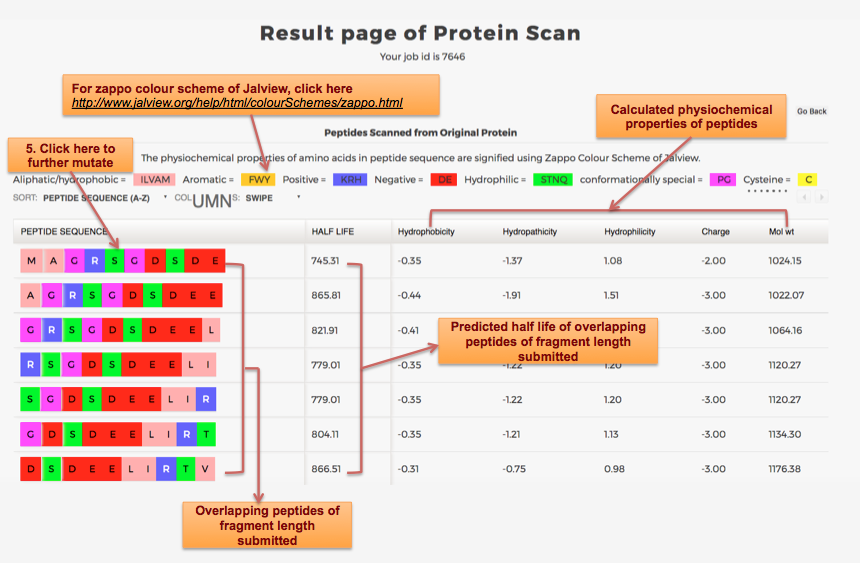
Designing of Half-life of Peptides from a Protein Sequence
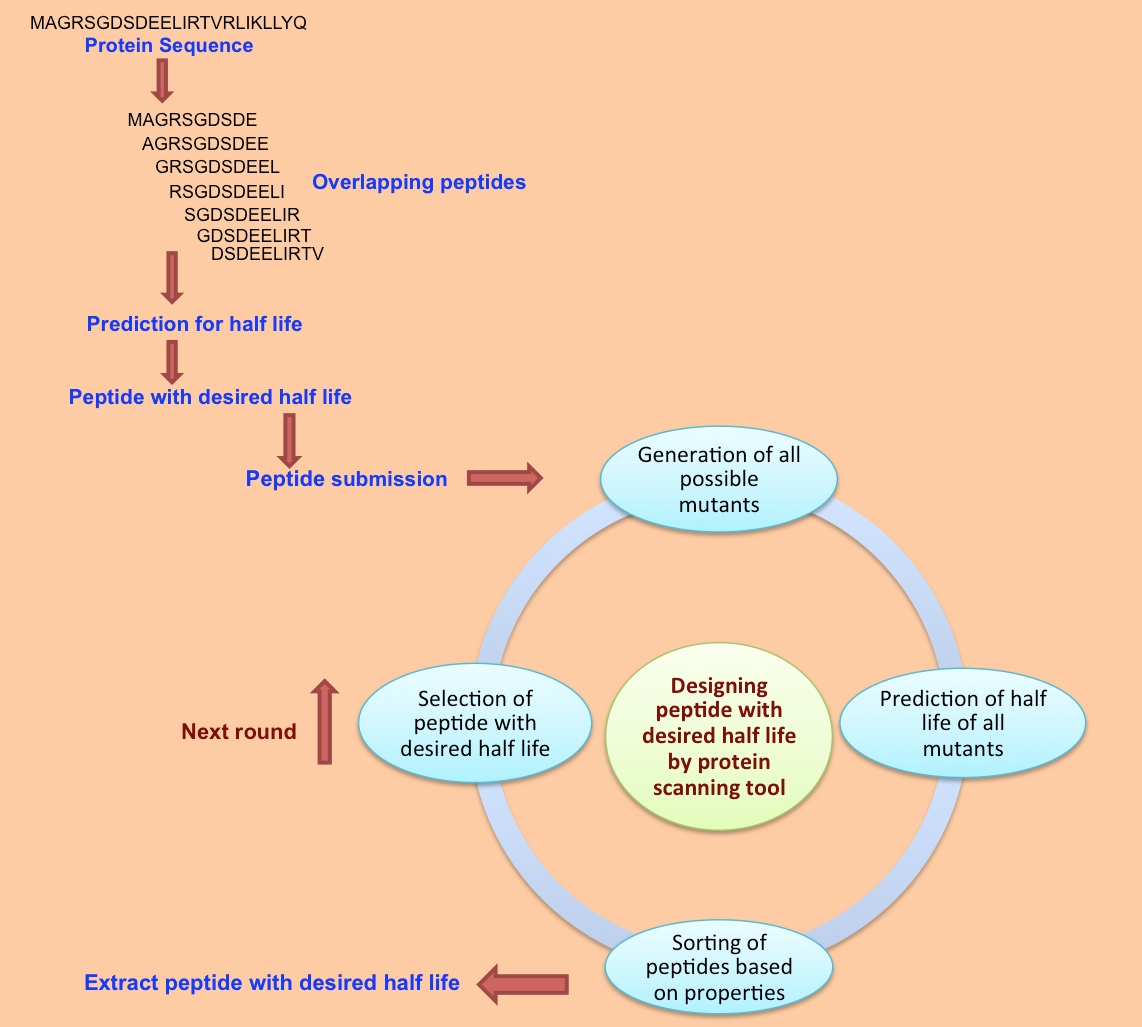
Example demonstrating use of Protein Scan for Designing Half-life of Peptides