|
Interlukin-6(IL-6) plays a vital role in the progression of novel coronavirus (COVID-19). Best of our knowledge no method has been developed for predicting epitope or antigenic peptides that can induce IL-6. First time an attempt have been made to develop method for predicting IL-6 inducing/non-inducing epitopes.
The prediction server for IL-6 inducing epitopes has been designed in a very user-friendly manner. Here, on this page, user can get the details of all the algorithms and procedures exploited in the different modules. |
|
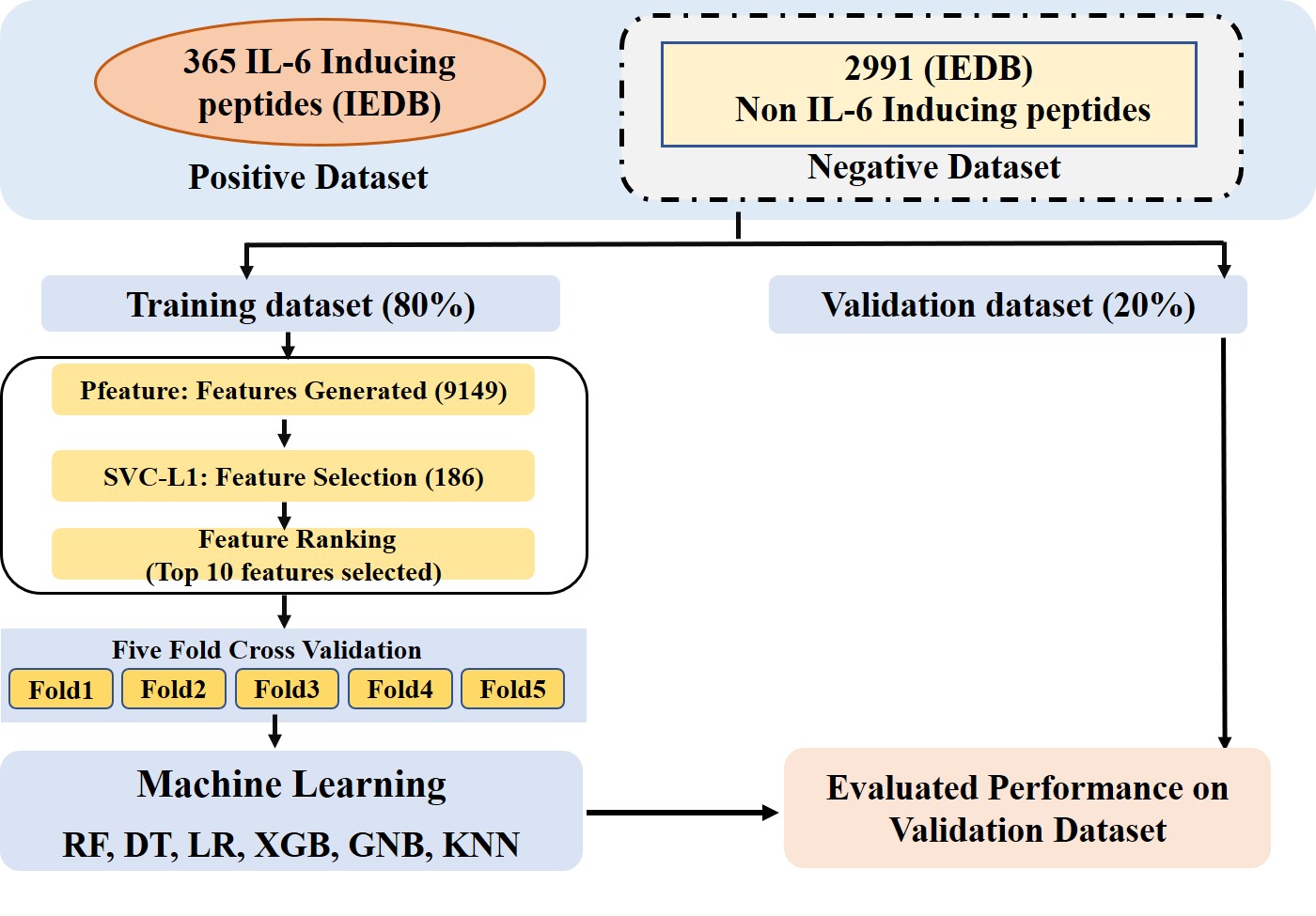
Dataset UsedPositive Dataset:It comprises of 365 experimentally validated IL-6 inducing peptides/epitopes.Negative dataset:It comprises of 2991 experimentally validated non-IL-6 inducing peptides/eptitopes . |
This algorithm of the server is relies on the following three models:
Prediction modelThe "Predict" module provides the facility to the user to classify IL-6 inducing peptides from non-inducing peptides. User can provide multiple sequences as input to the server to predict the given peptide is IL-6 inducing/non-inducing. In this study we used various machine learning techniques to develop prediction modules.Design modelIn this study, "Design" module is used to create all possible analogs/mutants of the input sequence and identify the best analog which initiate cytokine i.e. IL-6 release. Usign various machine learning techniques it can predict wheather mutants can induce IL-6 secretion.Scan modelIn this study, we incorporated three scan modulesI) Protein Scan: Aim of this approach is to identify IL-6 inducing regions in the protein sequence, which can futher removed or altered in the potential subunit vaccine candidates. 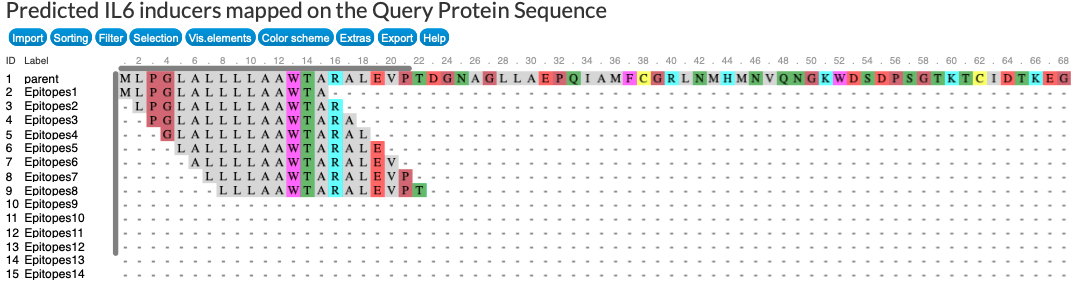
II) Motif Scan: Aim of this approach is to discover motifs or patterns in IL-6 inducing and non-inducing epitopes. In this study we used a powerful pattern discovery software MERCI. First we identified motifs are exclusively found in IL-6 inducing peptdes and not present in non-inducing peptides. Similarly we identified motifs in non-inducing peptides which are absent in IL-6 inducing peptides. These novel motifs used for discriminating IL-6 inducing and non-inducing peptides. III) BLAST Scan:“Blast Search” module is based on a similarity search method, i.e., Basic Local Alignment Search Tool (BLAST). The input query sequence is searched against the database of known IL-6 inducing peptides. A query sequence is predicted as IL-6 inducer if found match or hit in the database; otherwise, it is predicted as non-IL-6 inducer peptide. |