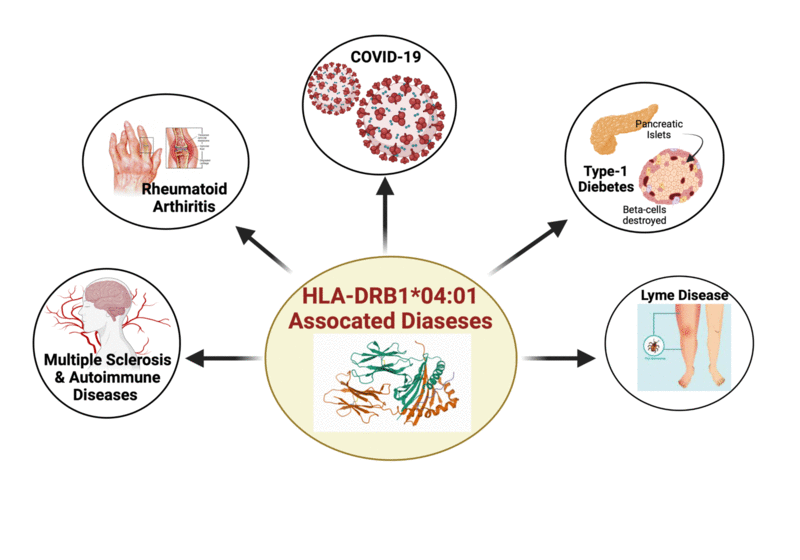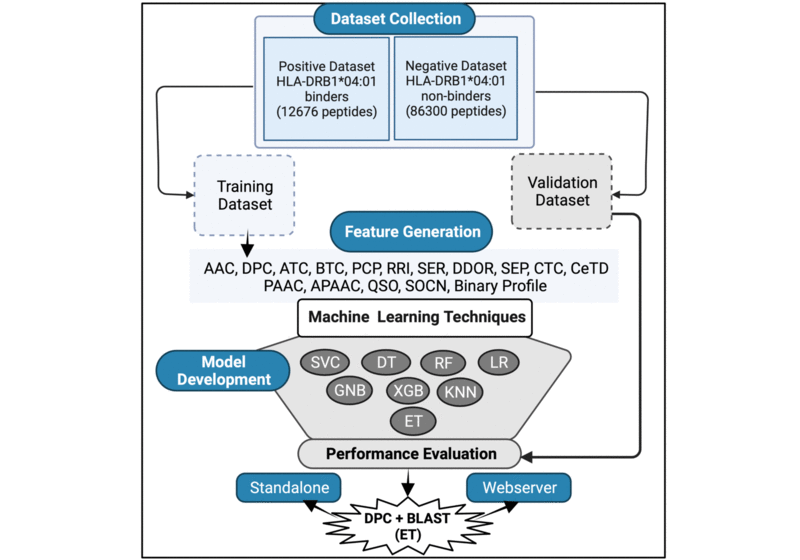
HLA-DR4Pred 2.0 is the update of the older version HLA-DR4Pred, which was a SVM and ANN based method to predict the HLA-DRB1*04:01 bindind peptides. In the updated version, the models were trained on a dataset comprising of 12676 HLA-DRB1*04:01 binders and 86300 non-binders. The performance of the models were evaluated using 5-fold cross-validation. The models were trained on dipeptide composition as well as on hybrid features(dipeptide composition + BLAST-search) by implementing extra-tree classifiers from the scikit library of python. The datasets were extracted from Immune Epitope Database (IEDB). This method will be highly useful in the fields of cellular immunology, immunodiagnostics, immunotherapeutics, and will aid in molecular understanding of autoimmune susceptibility.
References:
-
Patiyal S, Dhall A, Kumar N, Raghava GPS (2024) HLA-DR4Pred2: an improved method for predicting HLA-DRB1*04:01 binders. Methods, doi.org/10.1016/j.ymeth.2024.10.007.
-
Bhasin M, Raghava GPS (2004)SVM based method for predicting HLA-DRB1*04:01 binding peptides in an antigen sequence. Bioinformatics 20(3): 421-3