Welcome to the Help Page of HLA-DR4Pred2.0
This webserver has been designed to help the scientific community in order to predict, scan, or design the HLA-Class II allele HLA-DRB1*04:01 binders using sequence information. There are five mojor modules available in the server such as: |
This page provides help on different modules of server. Following are major modules in this server:
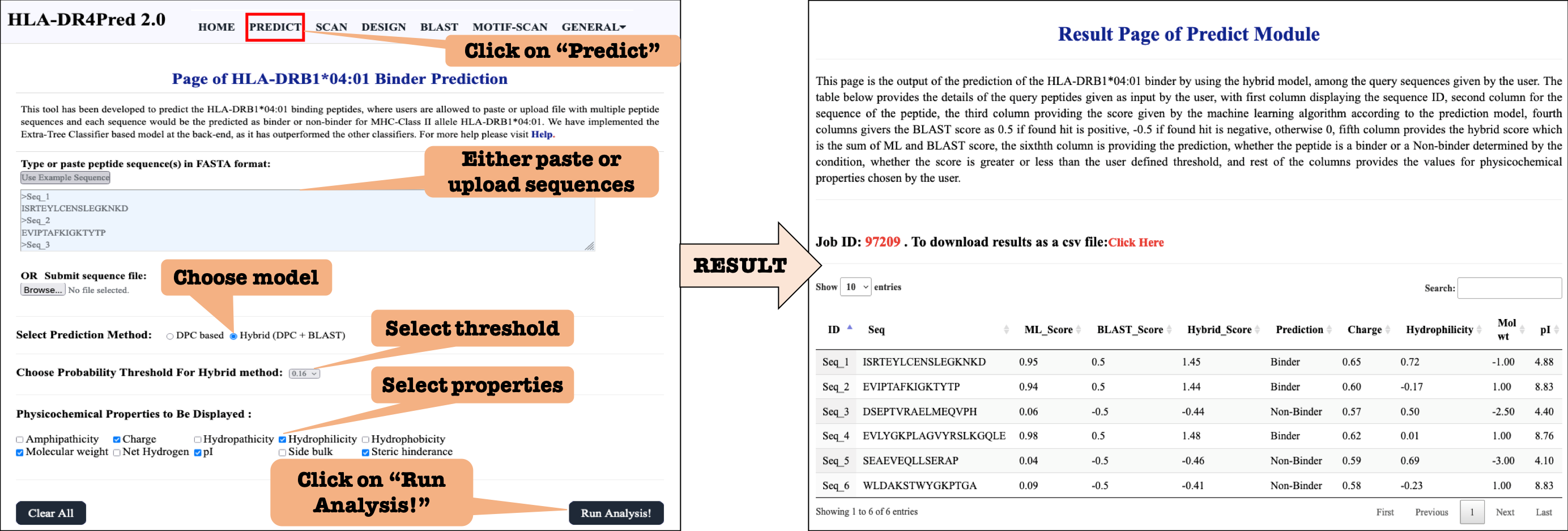
This module allows users to predict the HLA-DRB1*04:01 binding peptide in a set of peptides or peptide library (virtual screening). User may submit large number of peptides and server will predict and rank them based on their potential to bind HLA-DRB1*04:01.
PREDICT:
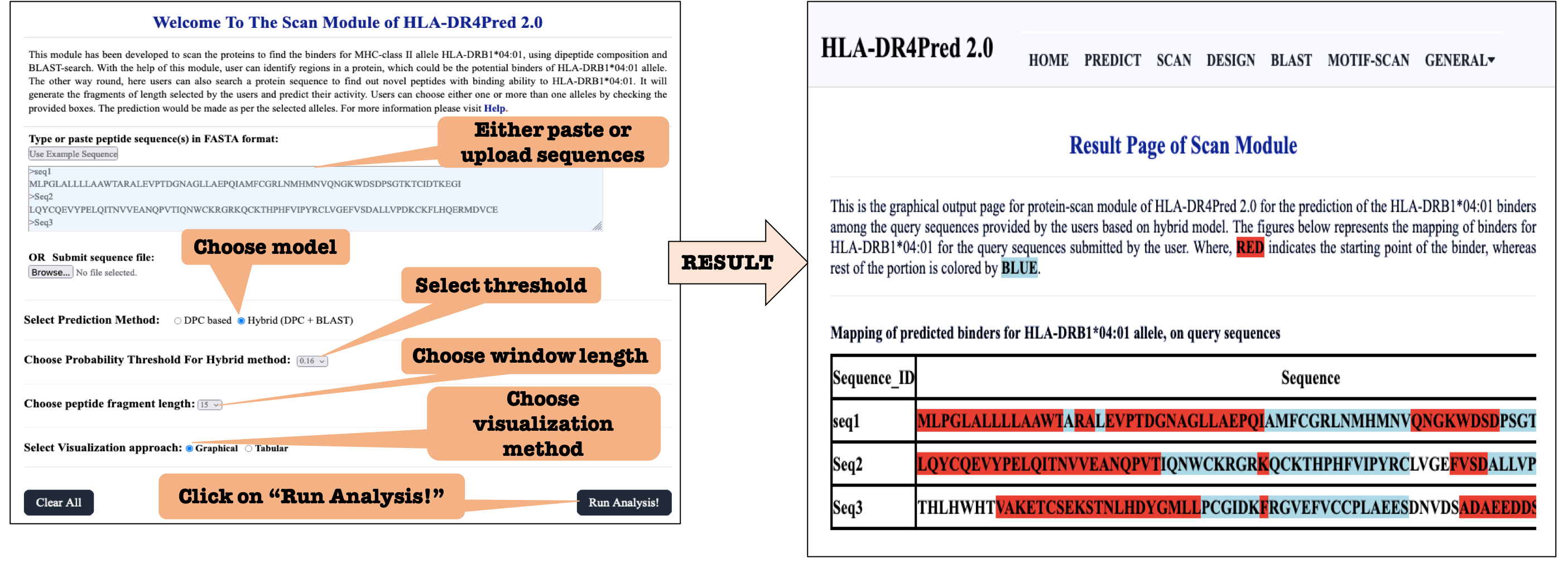
SCAN: This module allows user to identify regions in a protein, which can act as HLA-DRB1*04:01 binders, which may be further removed or altered. The other way round, here users can also search a protein sequence to find out novel HLA-DRB1*04:01 binding peptides.
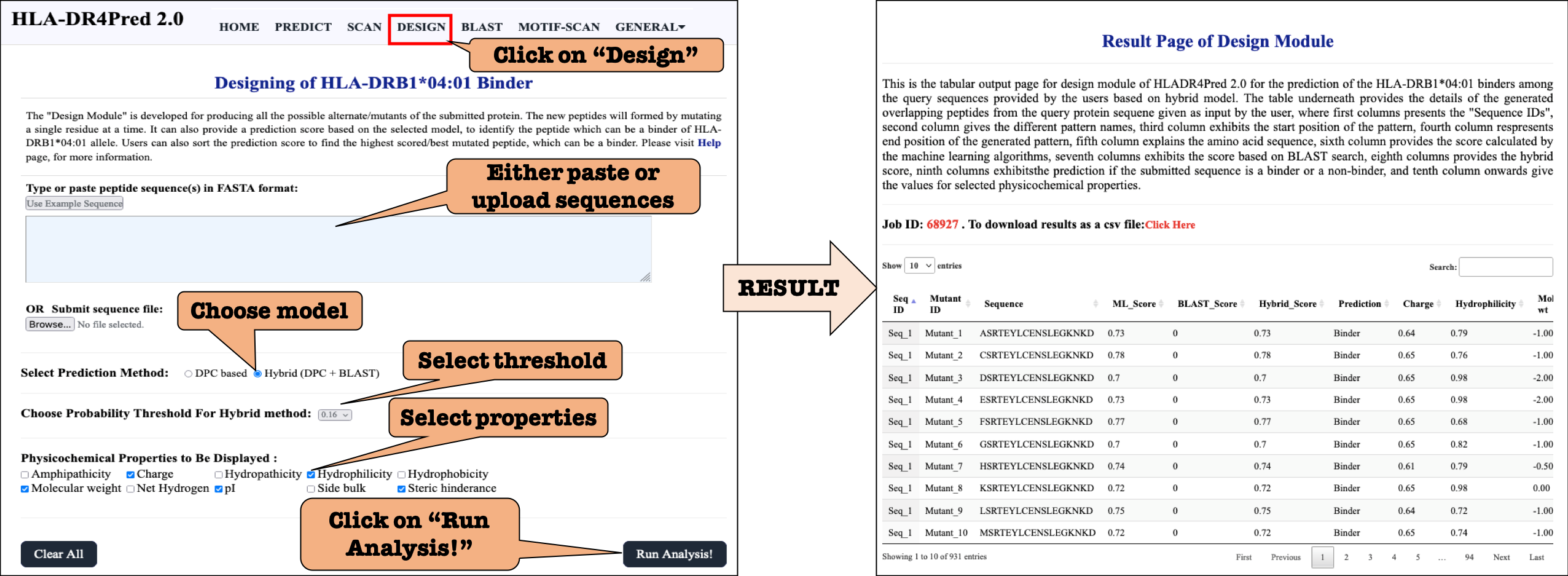
DESIGN: It generate all the plausible mutants of the query peptide(s) and predict HLA-DRB1*04:01 binders in mutants. This server allows users to identify minimum mutations required to increase its potency.
BLAST: This page allows users to search the query peptide sequence(s) against the database of known HLA-DRB1*04:01 binders using similarity-based search method, i.e. BLAST.

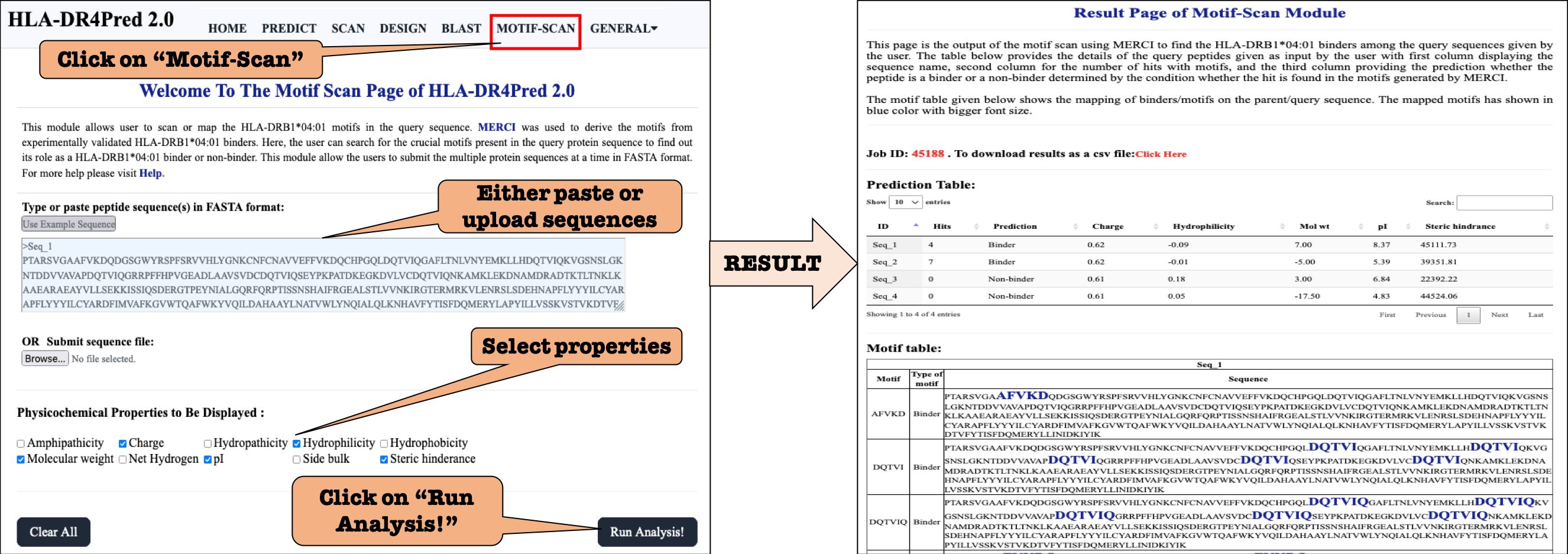
MOTIF-SCAN: This page allow users to search for motifs derived using known HLA-DRB1*04:01 binders, in the query peptide sequence(s) using MERCI software.