Help page of CDpred
The help page provides the detailed instruction on how to use the CDpred webserver and its modules. The various CDpred modules described below were used to build the architecture of this help page.
Major modules of CDpred
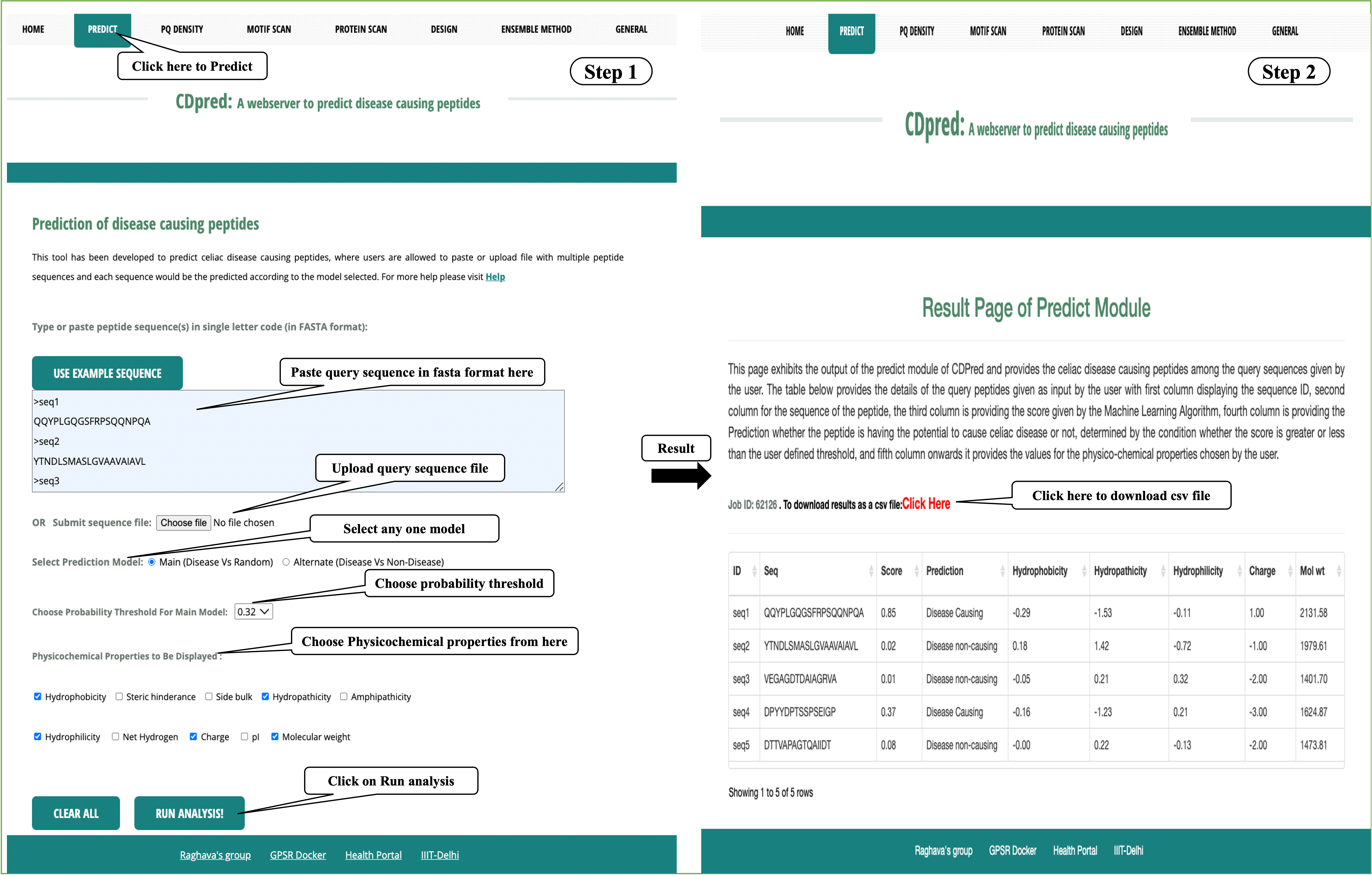
Predict: This module allows users to predict the celiac disease causing peptide/epitope in a set of peptides. User may submit large number of peptides and server will predict peptide sequences according to selected model.
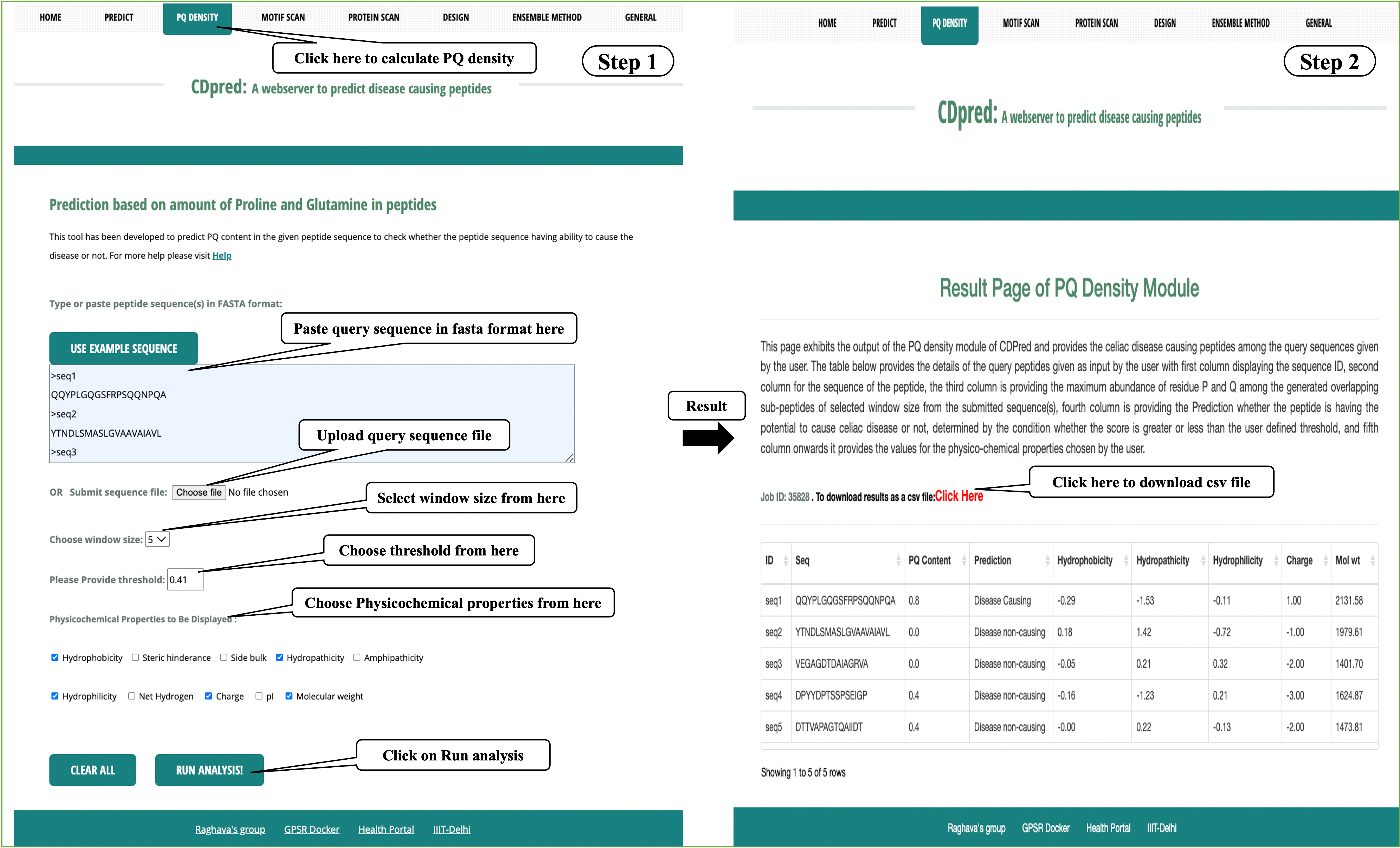
PQ Density:
This module helps to predict the composition of PQ residues in the given peptide sequence. User may submit large number of peptides and server will calculate the omposition of PQ residues by generating different overlapping windows.
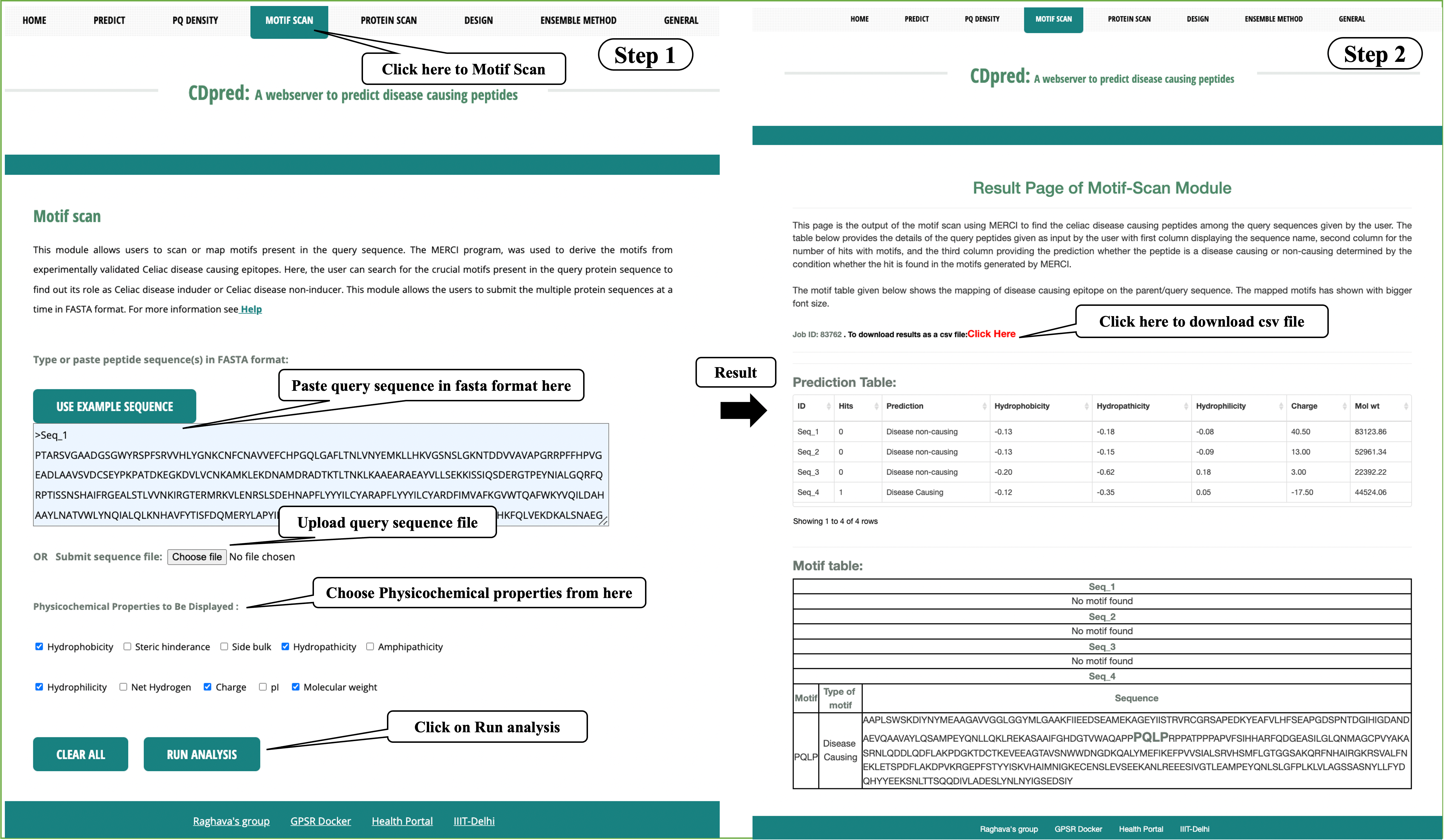
Motif Scan: This module allows users to scan or map celiac disease causing motifs which may be present in the query sequence, using MERCI program.
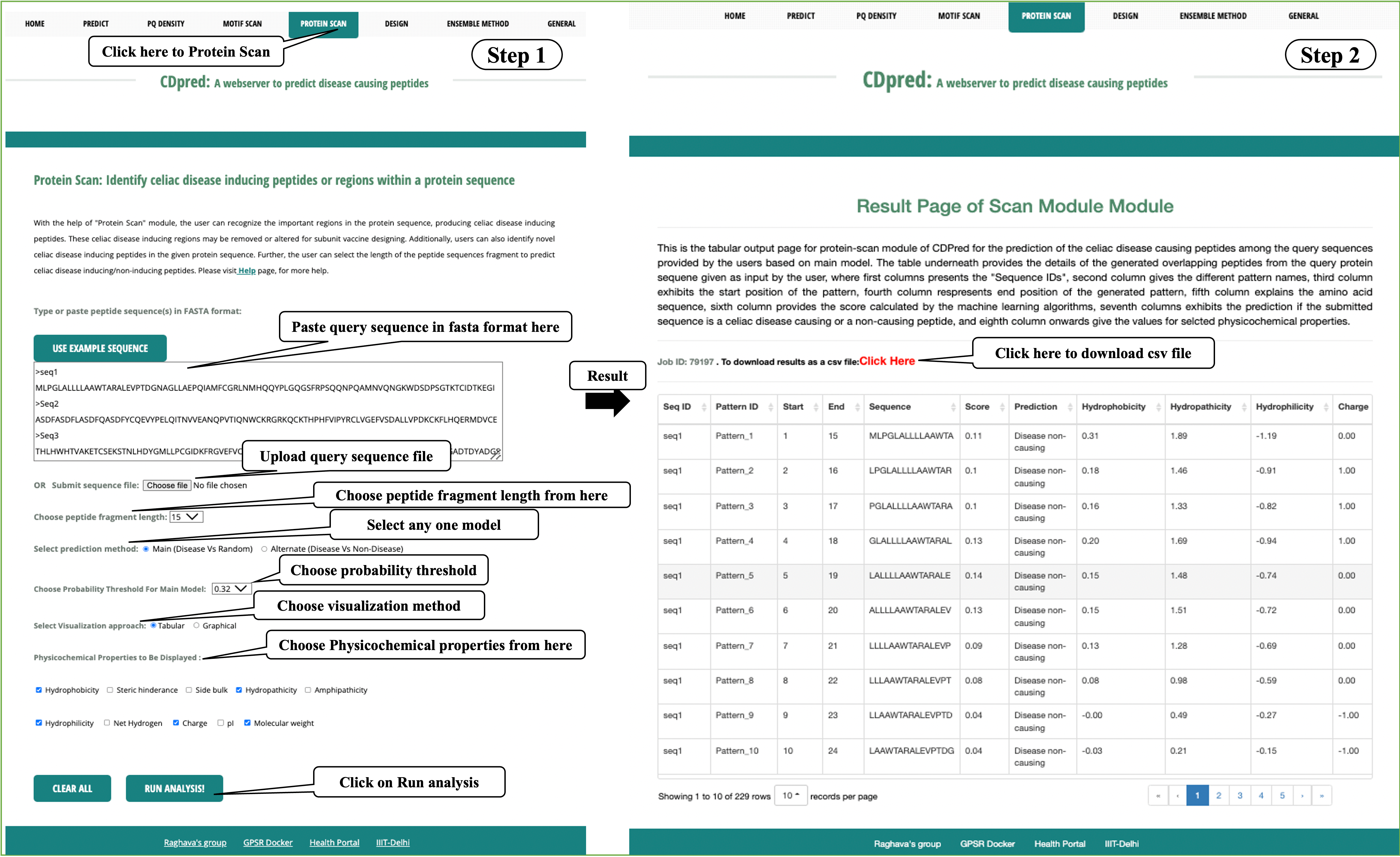
Protein Scan: This module allows user to identify regions in a protein, which are contributing in the development of celiac disease, which may be further removed or altered. The other way round, here users can also search a protein sequence to find out novel celiac disease causing peptides.
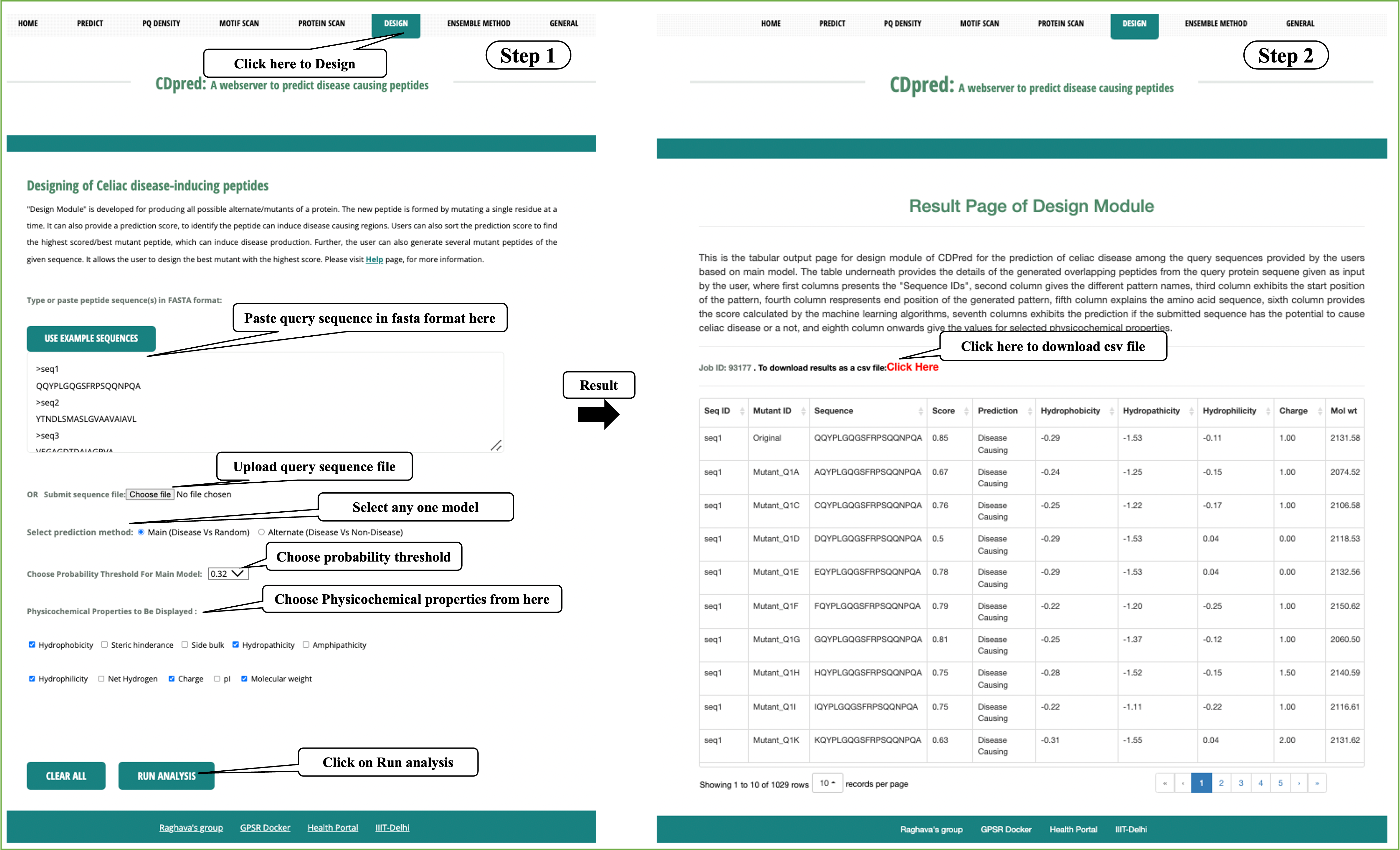
Design: It generate mutant of a query peptide and predict celiac disease causing epitopes in mutants. This server allows users to identify minimum mutations required to increase its potency.
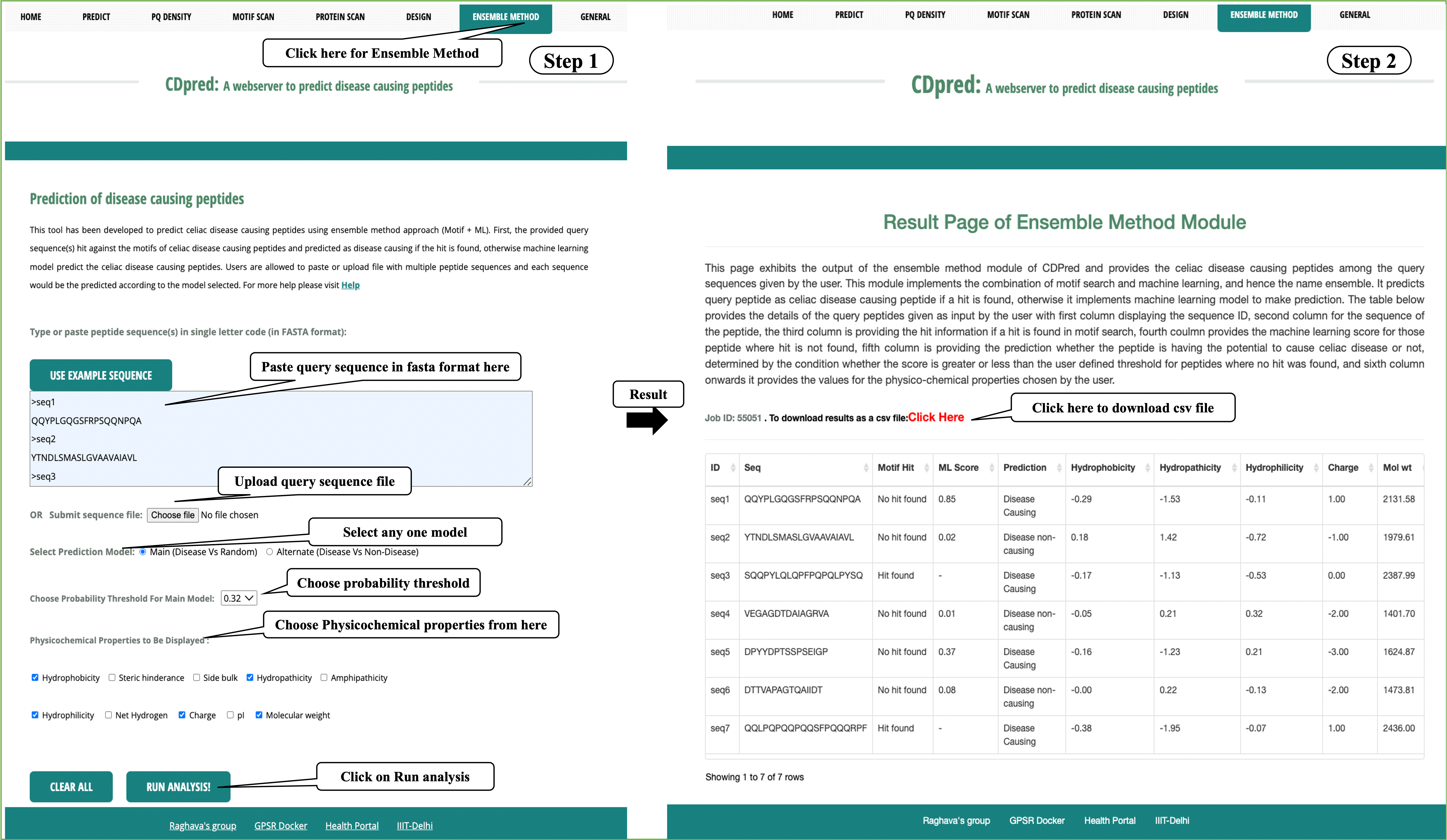
Ensemble Method:
This module helps to predict the disease causing peptides using ensemble method approach (Motif and ML based). The module use both motif based approach as well as machine learning models for the prediction.