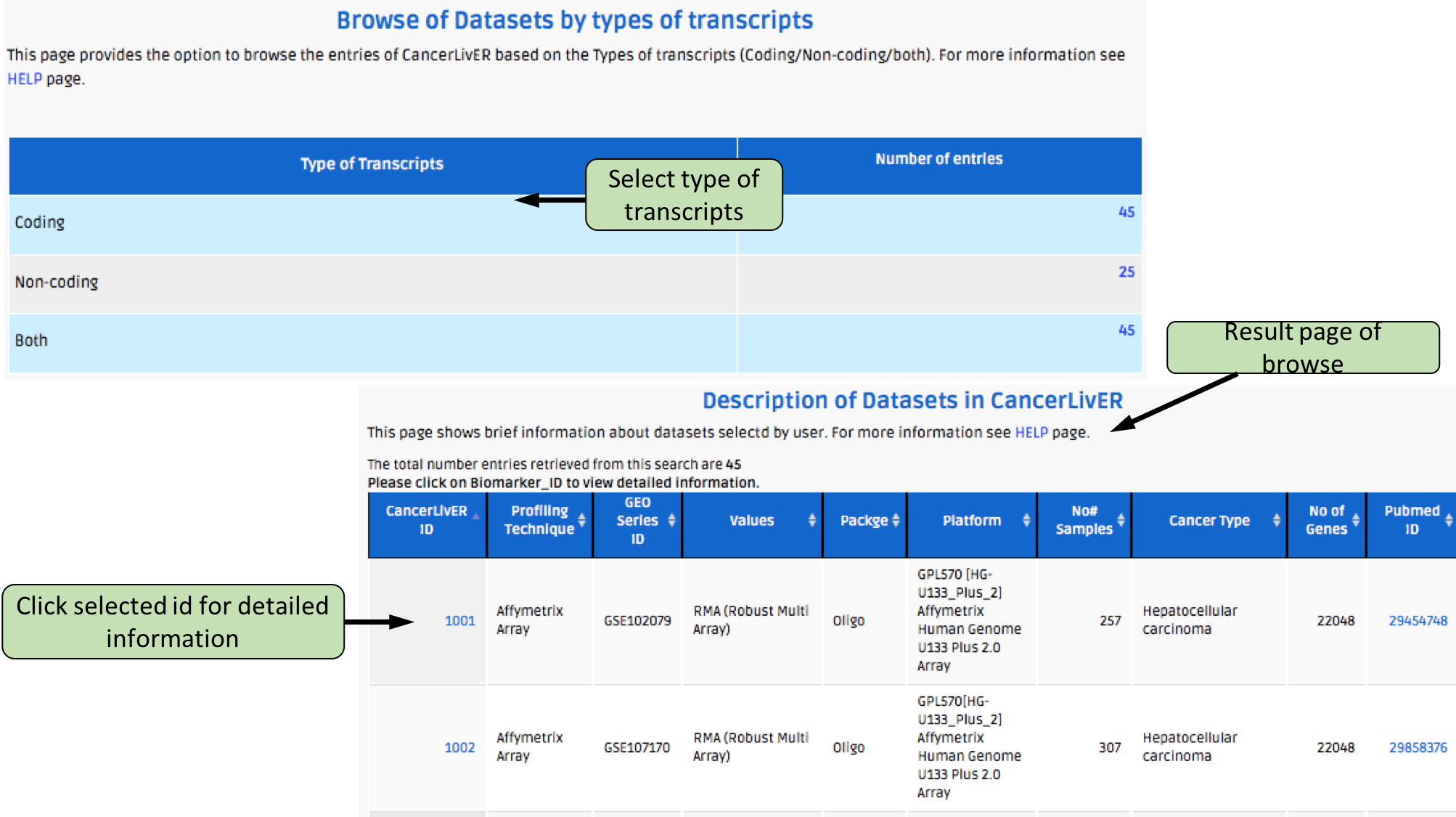
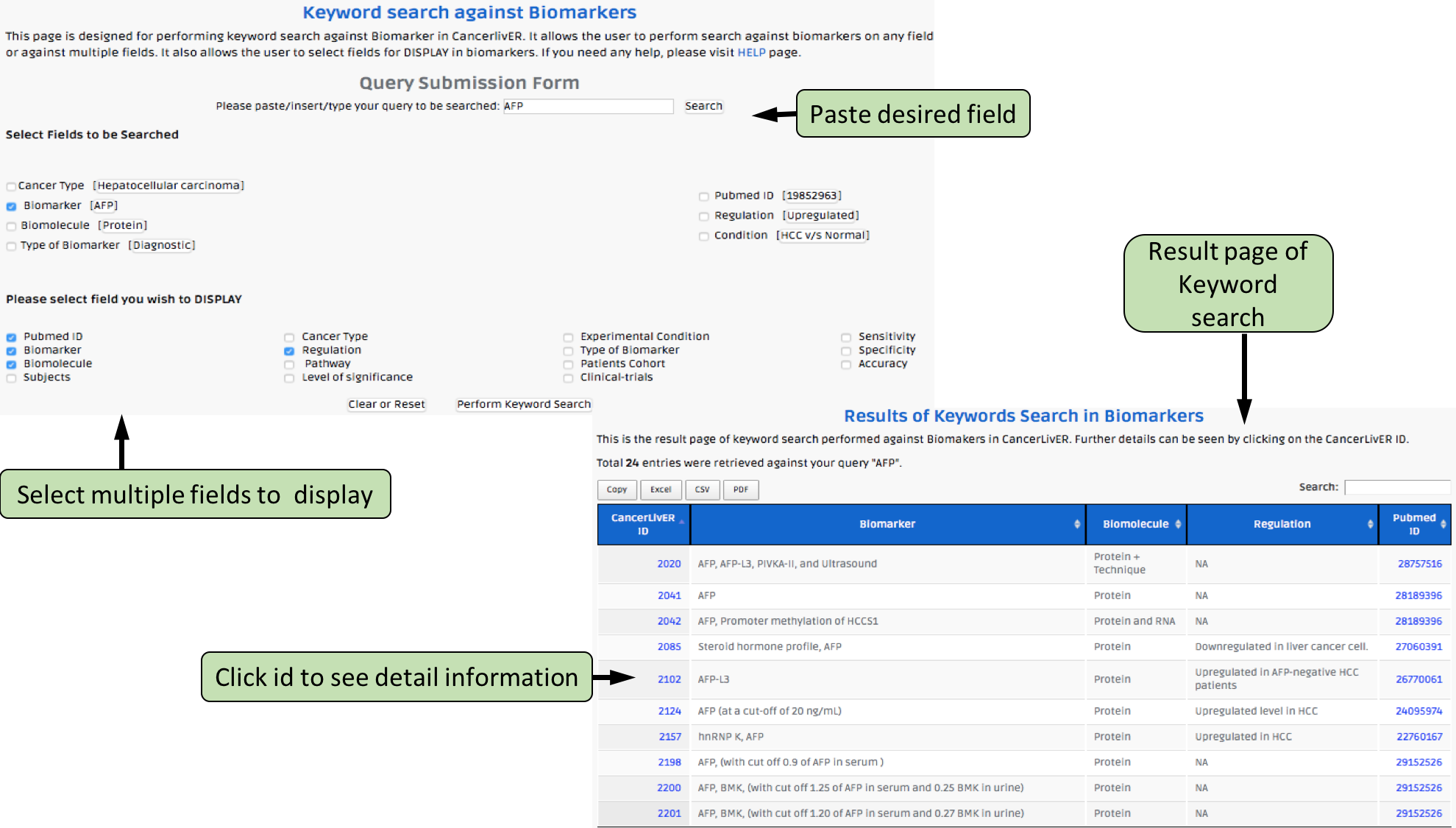
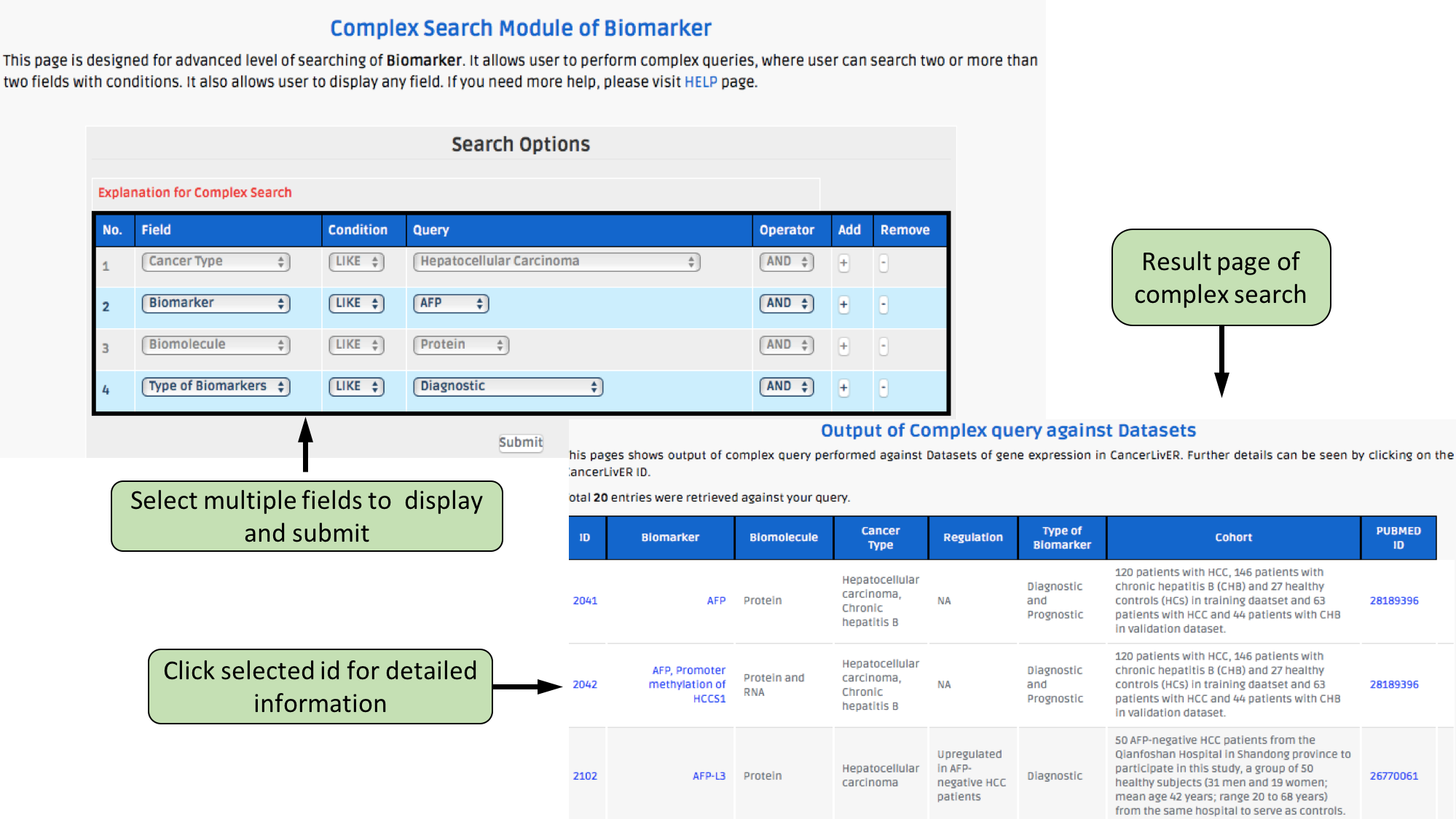
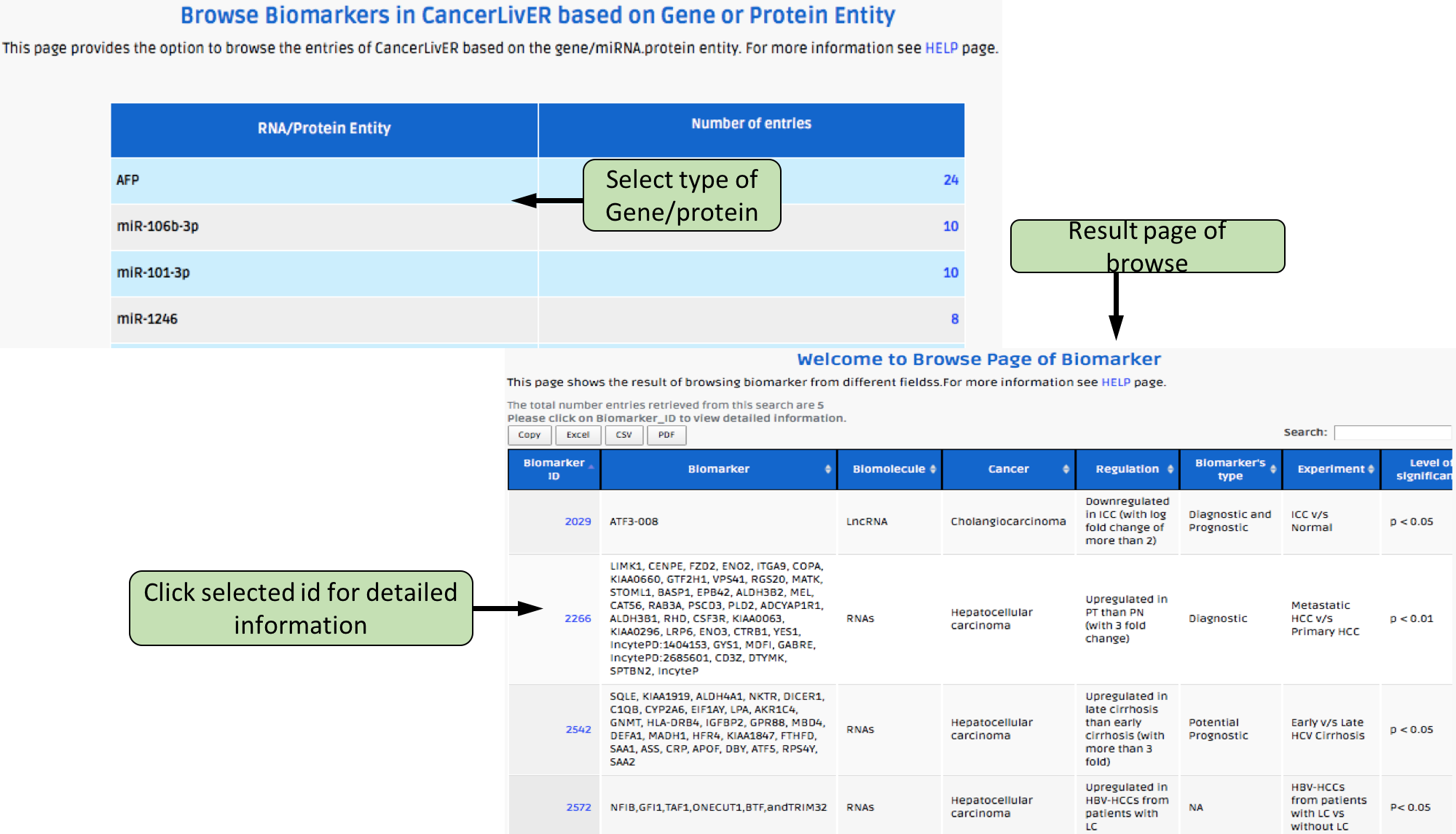
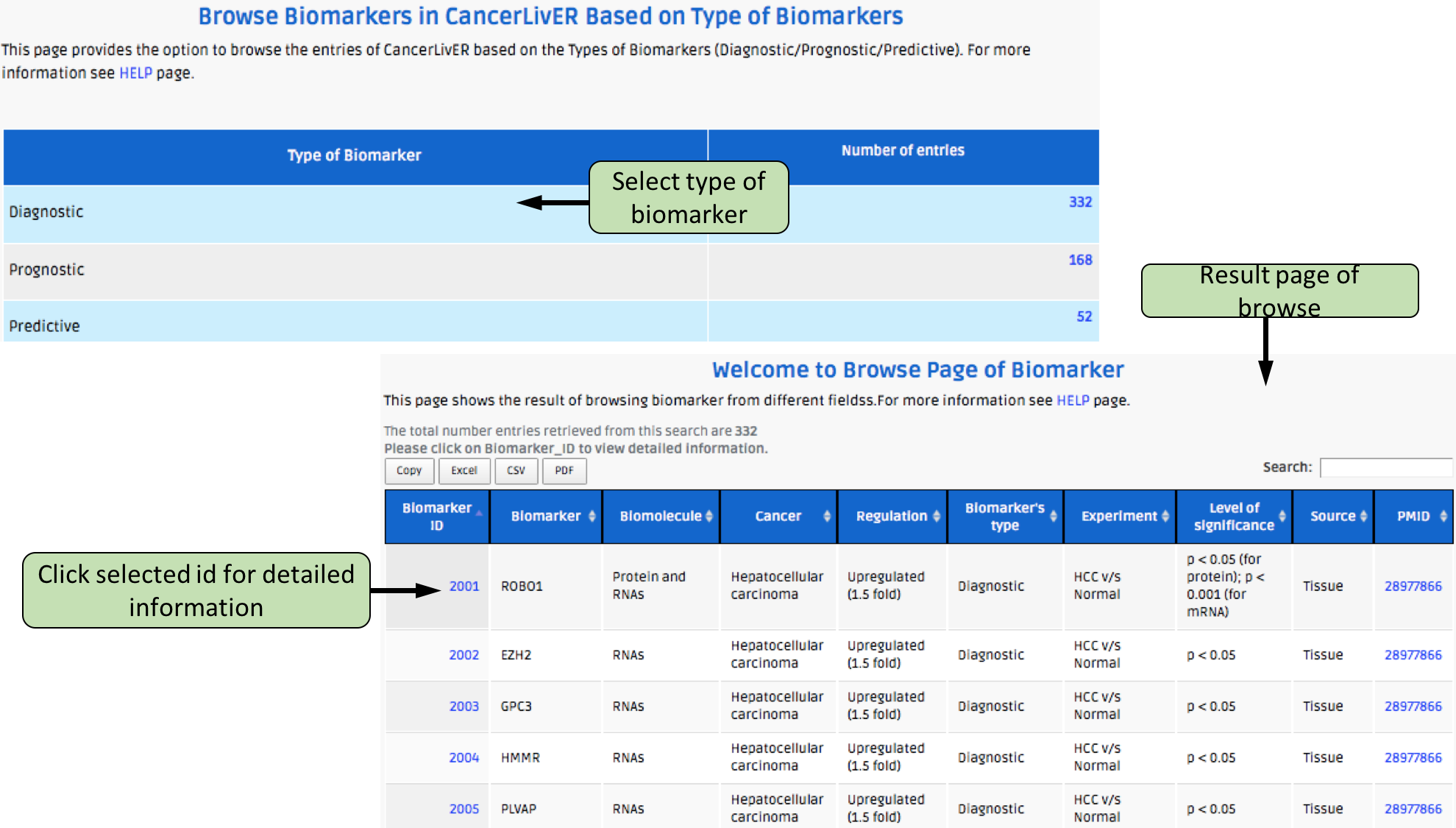
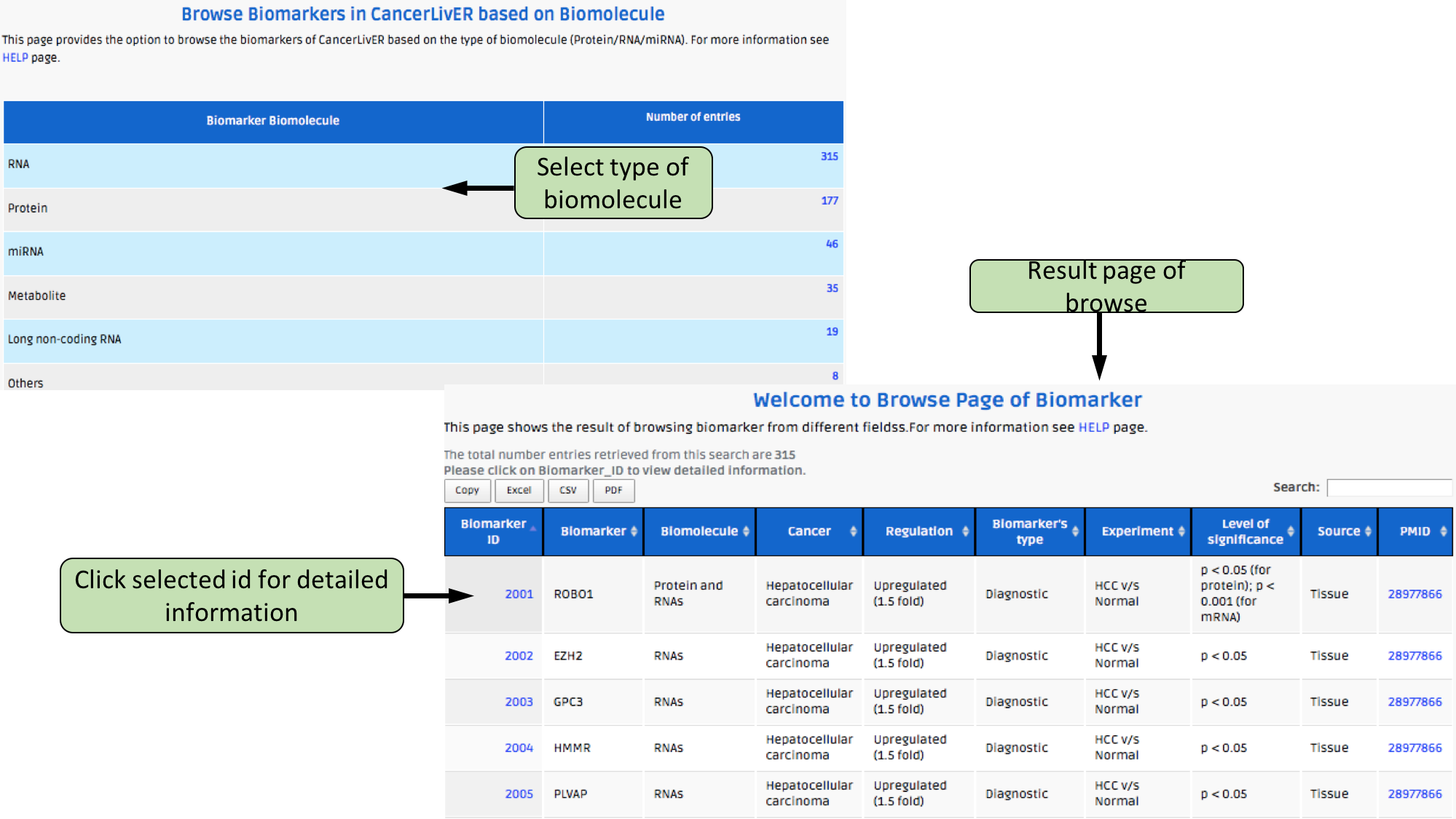
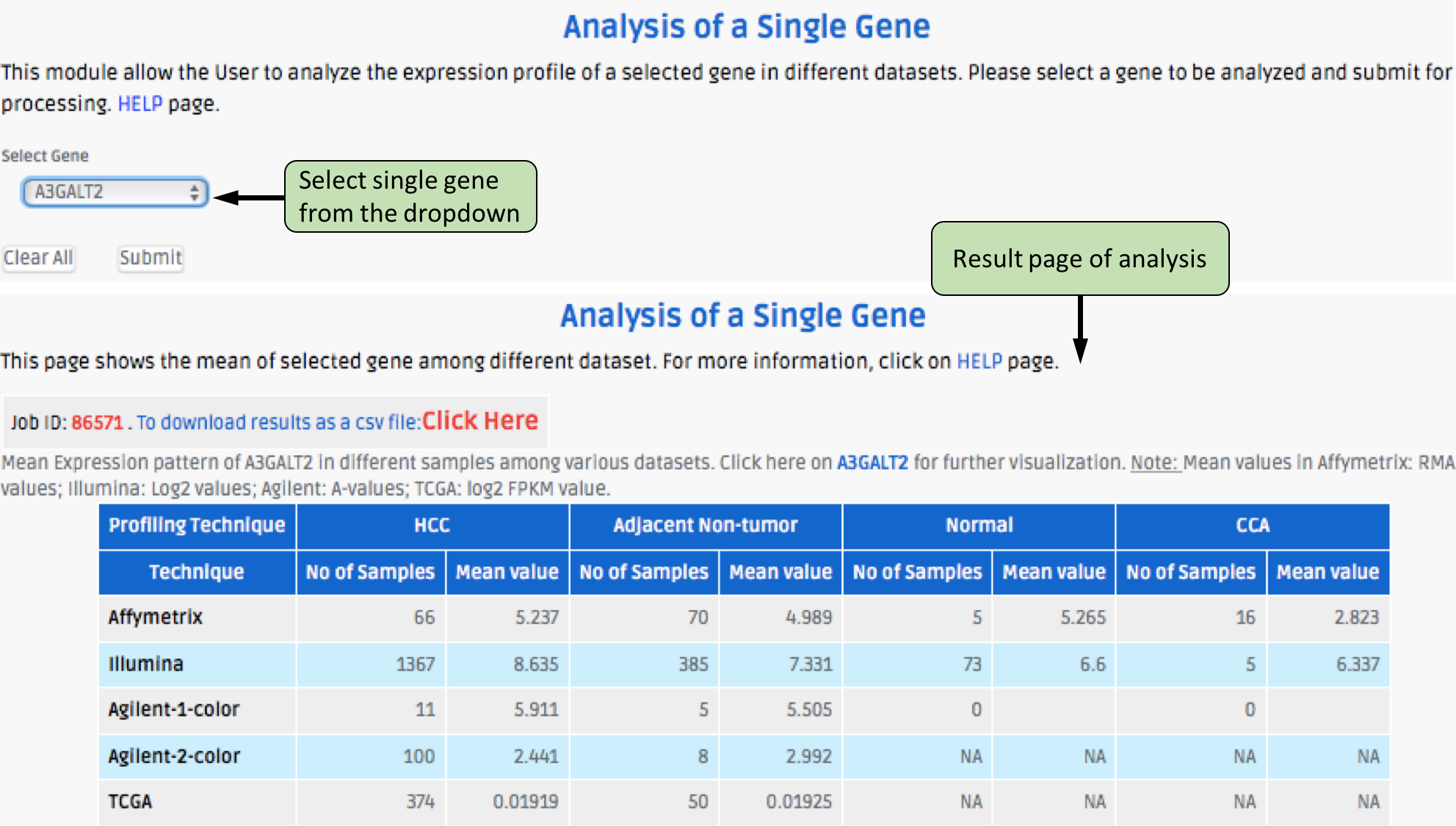
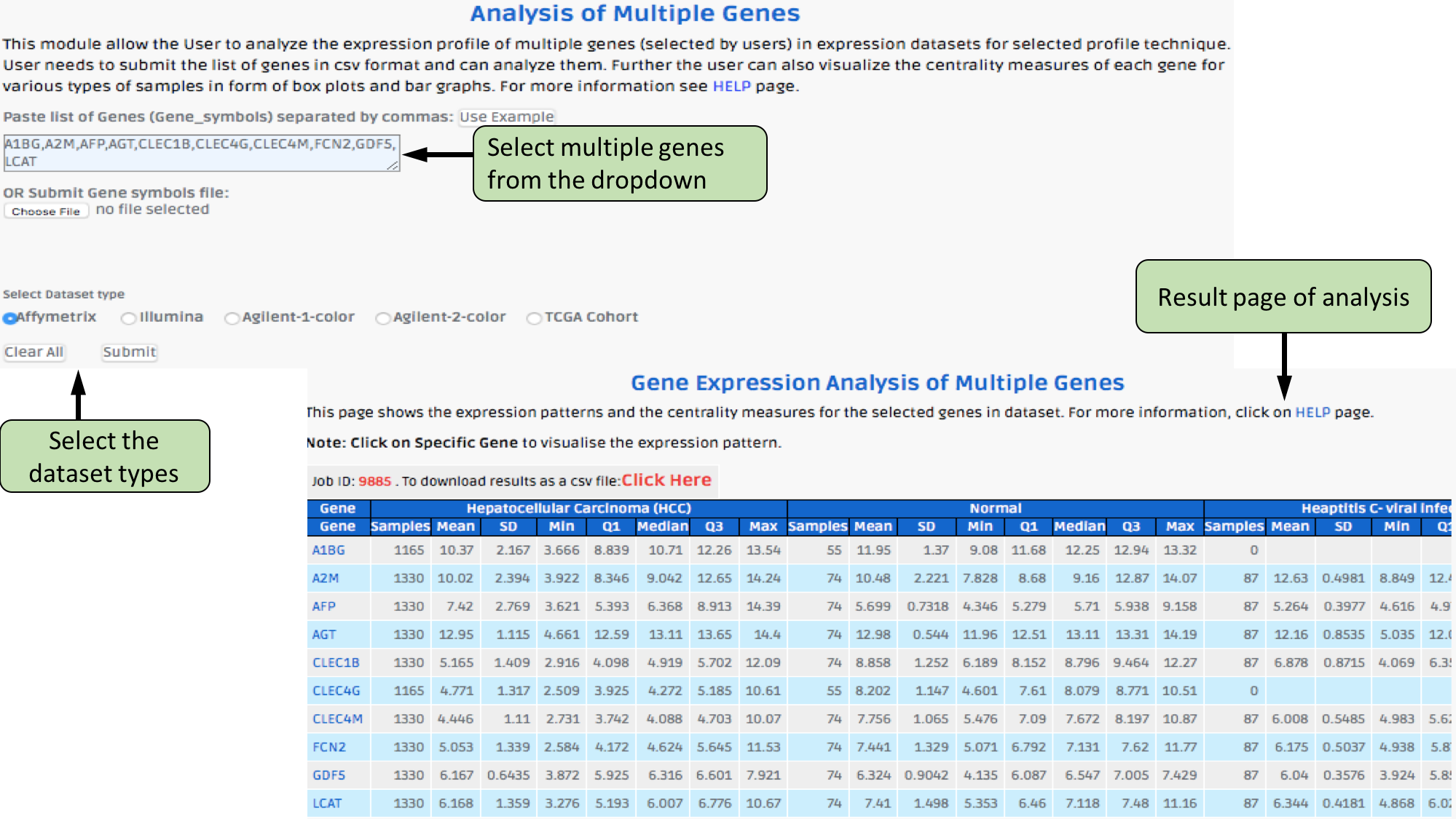
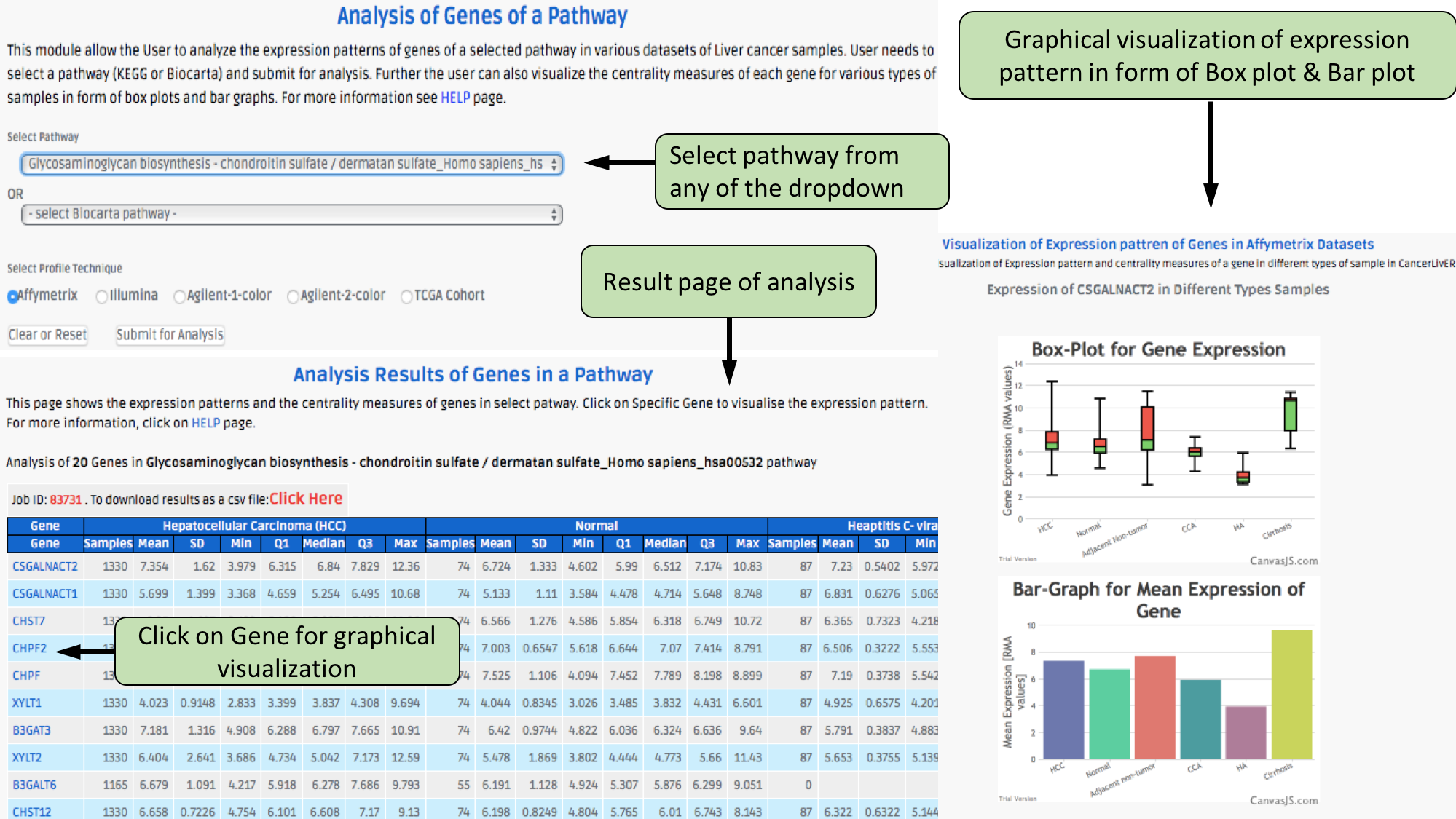
Complex Search
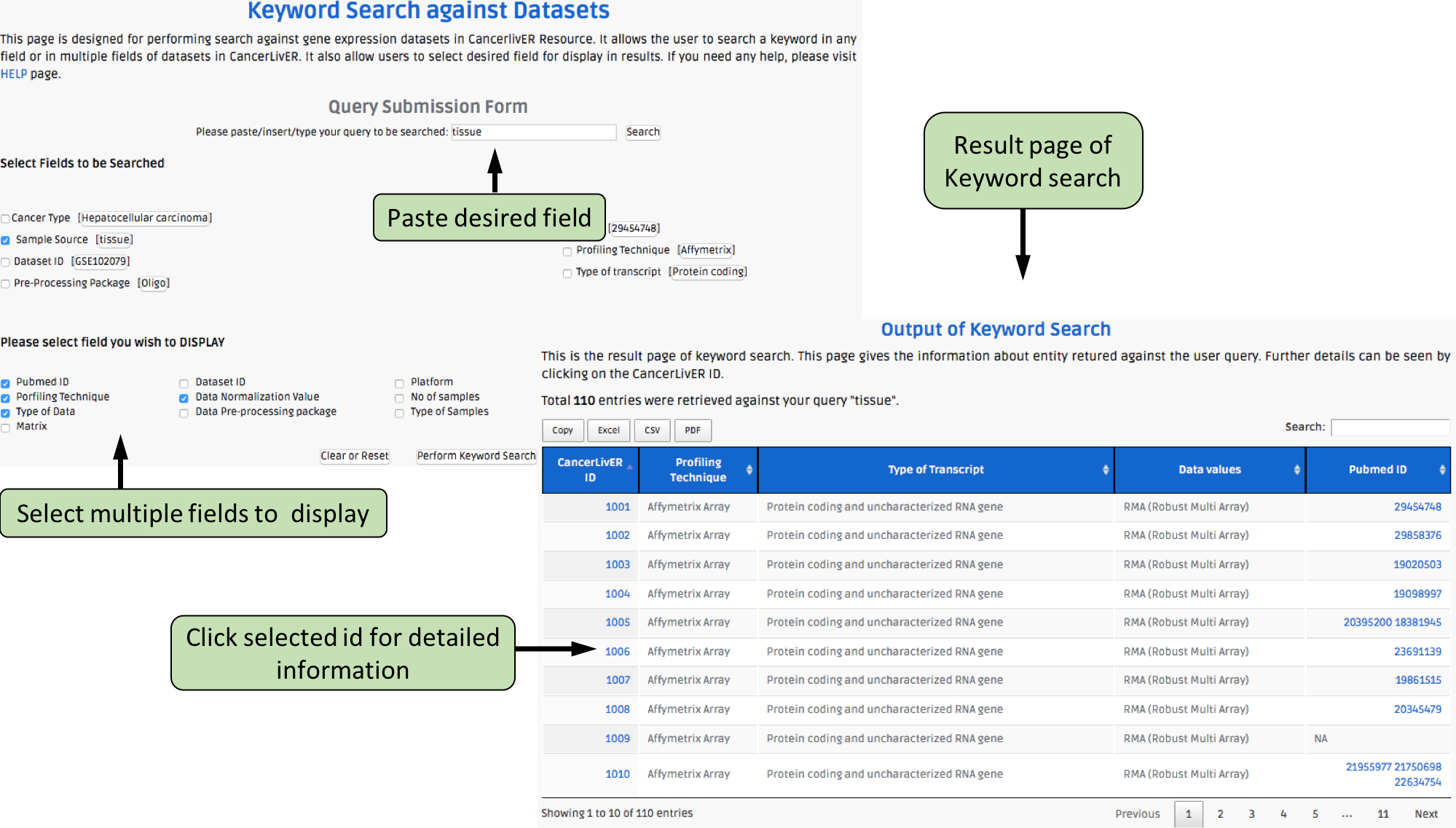
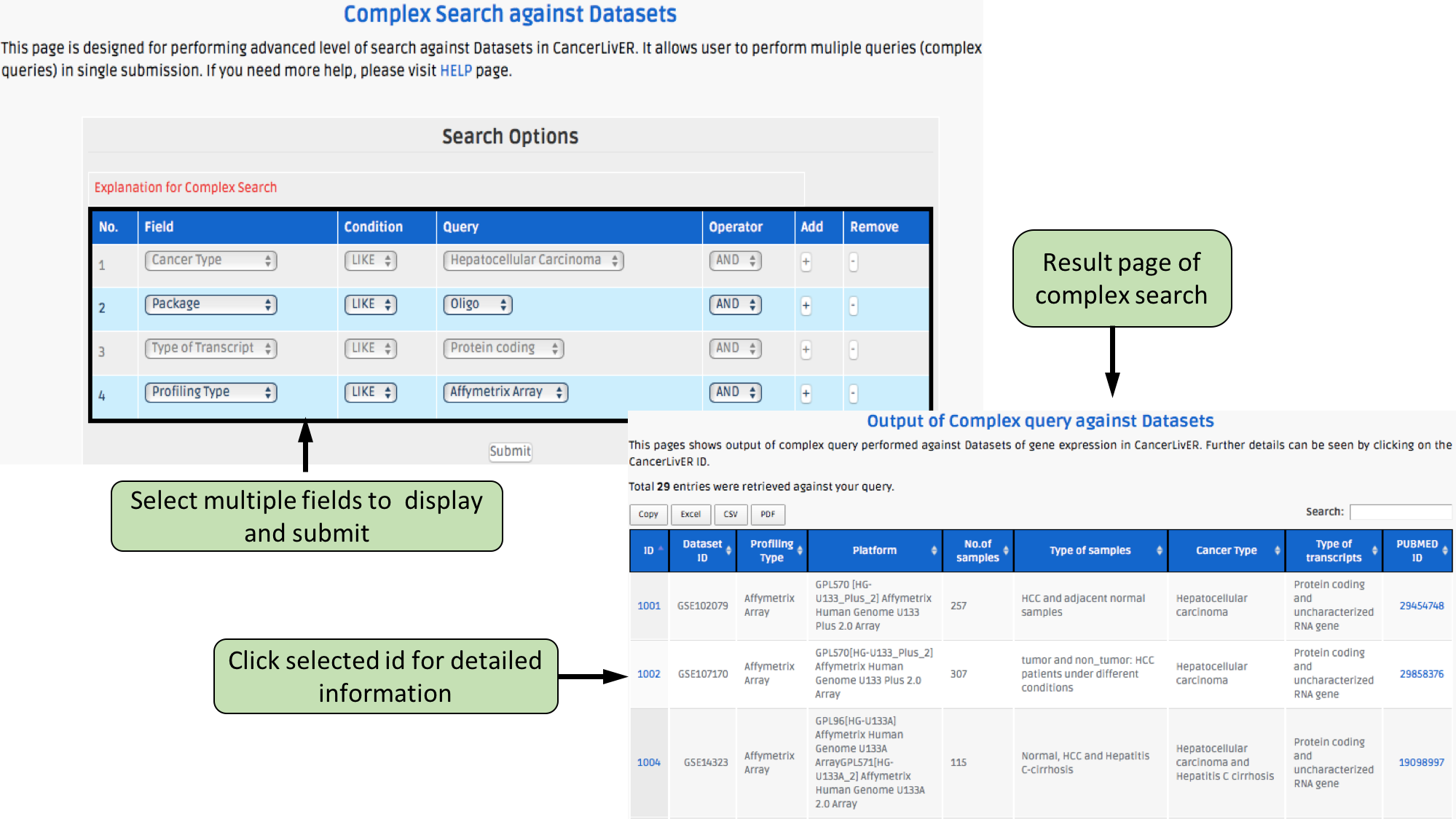
This page provides the advanced search facility in CancerLivER. User can browse the page by selecting two or more than two fileds.
|
| Complex Search is quite helpful when user has predefined query in mind. For instance, User come up with the following questions in mind and wants to SEARCH |
| 1.Select cancer type like Hepatocellular carcinoma. |
| 2. Among the above selected cancer type select package like oligo. |
| 3. Among the above selection criteria, show type of trancript like protein coding. |
| 4. Among the above, select profiling technique like affymetrix array. |
| By clicking Submit; user can get desired output. |
| In a similar way, query can be extended using "+" button and reduced using "-" button. |
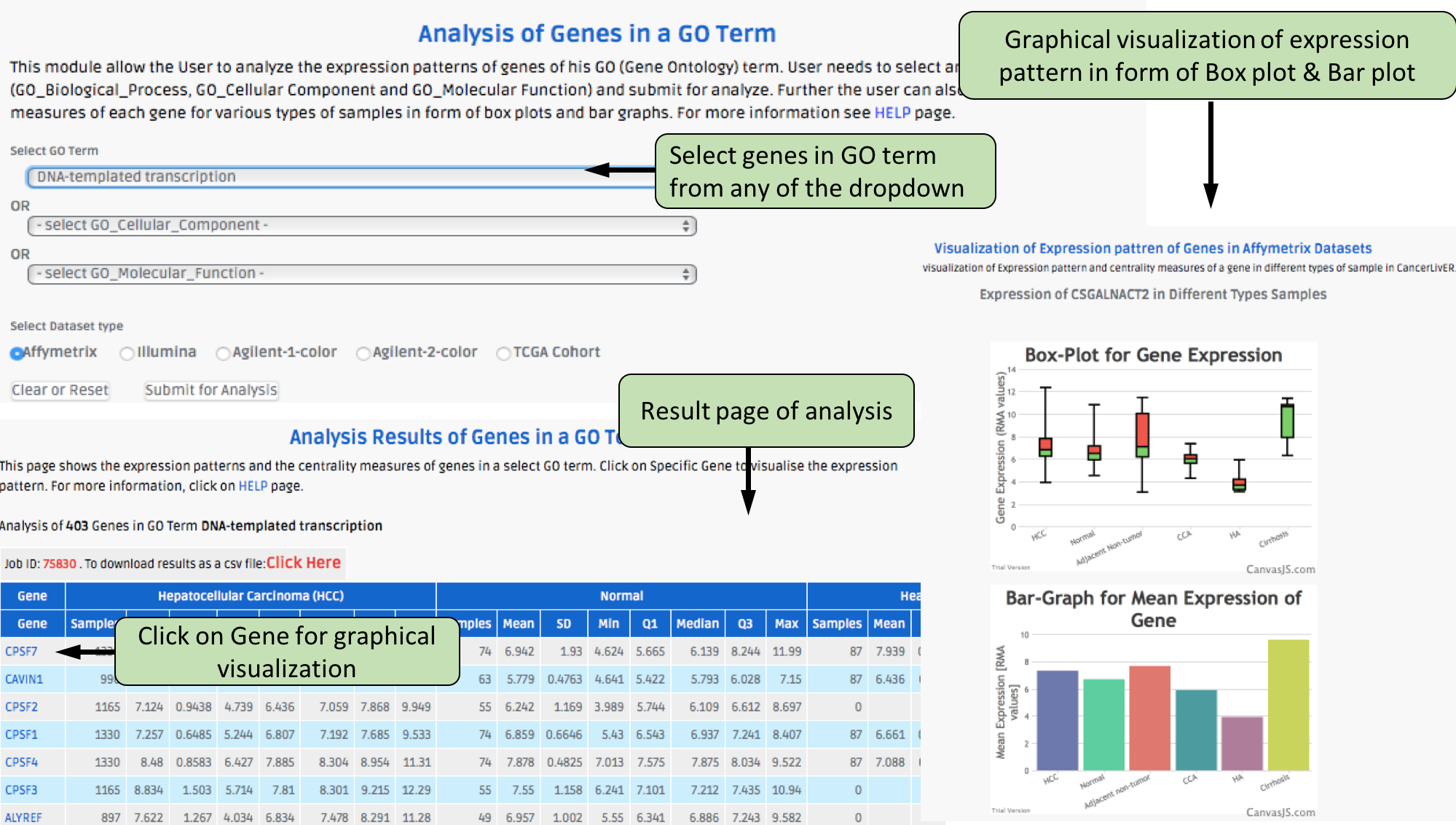
Complex Search
This page provides the advanced search facility in CancerLivER. User can browse the page by selecting two or more than two fileds.
|
| Complex Search is quite helpful when user has predefined query in mind. For instance, User come up with the following questions in mind and wants to SEARCH |
| 1.Select cancer type like Hepatocellular carcinoma. |
| 2. Among the above selected cancer type select package like oligo. |
| 3. Among the above selection criteria, show type of trancript like protein coding. |
| 4. Among the above, select profiling technique like affymetrix array. |
| By clicking Submit; user can get desired output. |
| In a similar way, query can be extended using "+" button and reduced using "-" button. |