About CancerDP |
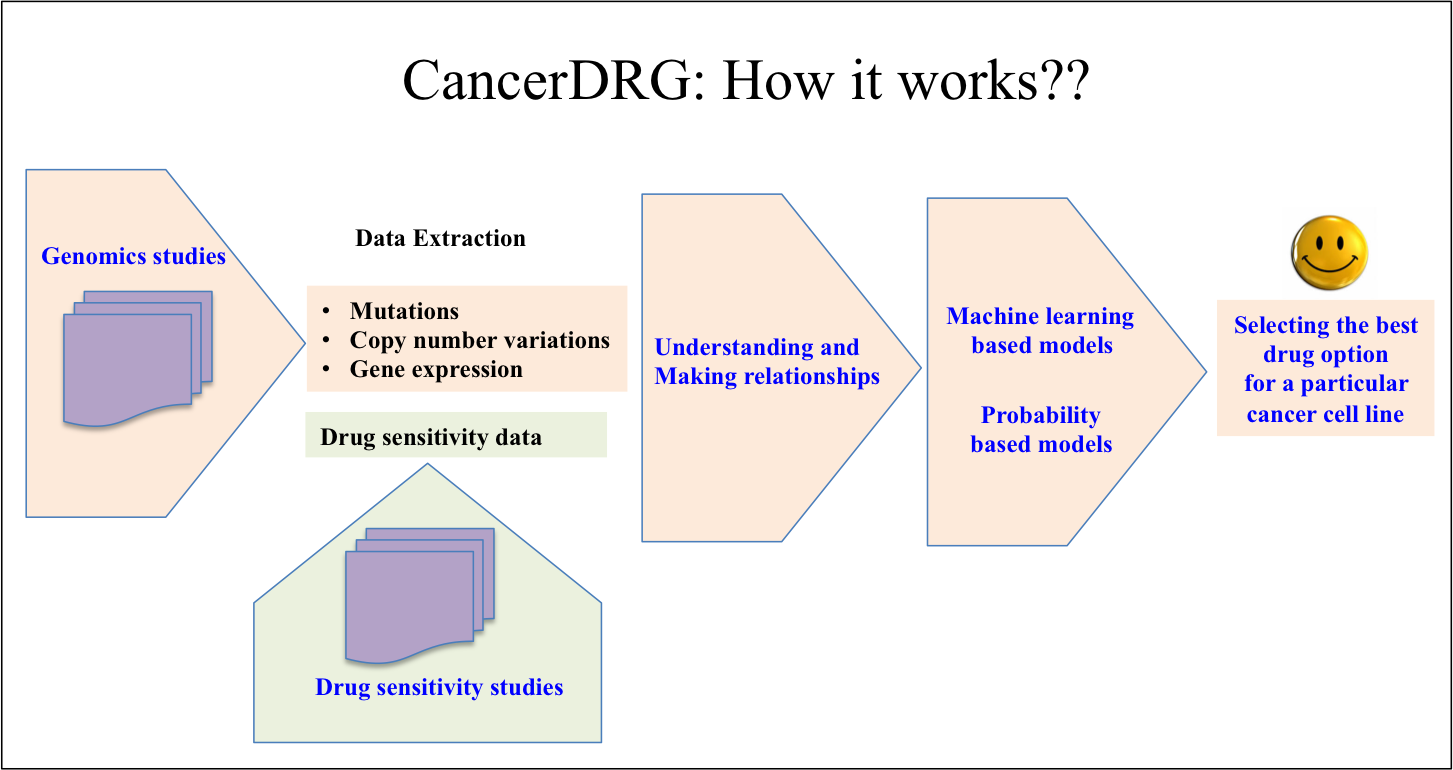
|
|
The aim of personalized treatment is to ascertain the response of specific cancer to a particular therapy. In the era of high throughput screening and next generation sequencing, this aim can be achieved by making relationships between genomic alterations and drug responses. With the advances in next-generation sequencing, thousands of genomes of almost all types of cancers have been sequenced, which provides valuable insights related to genomic alterations responsible for progression of the various cancers. At the same time, high throughput screening of hundreds of drugs against these cancer cell lines enable researchers the real response of a particular drug against cancer cell lines. Such data, if analyzed systematically, can be used to understand what will exactly happen if a particular therapy will be used against a particular cancer. Therefore, cancerDRG is an attempt in this direction to understand the relationships between genomic alteration and drug sensitivity against various cancer cell lines. In CancerDP, we have developed various in silico models using machine learning techniques, which will be helpful to select the best possible drug options against a particular cancer. For this, we have used the genomic and drug sensitivity information from CCLE. The genomic data includes sequencing of 1650 important genes, expression of 16582 genes and copy number variation of 16582 genes. The drug sensitivity data includes IC50 (μM) for 24 anti-cancerous drugs on >500 cell lines. Out of these 24 anti-cancerous drugs, 9 drug are launched, 2 are in phase III trial, 3 are in phase II trial and 7 are in preclinical trial. |
