Algorithm
The prediction server for predicting the protein as toxic and non-toxic has been designed in a very user-friendly manner. This page explains the compatibility of webserver on different operating systems, details of all the algorithms and methods used to develop models for prediction. Help Page
ToxinPred2 Methodology:
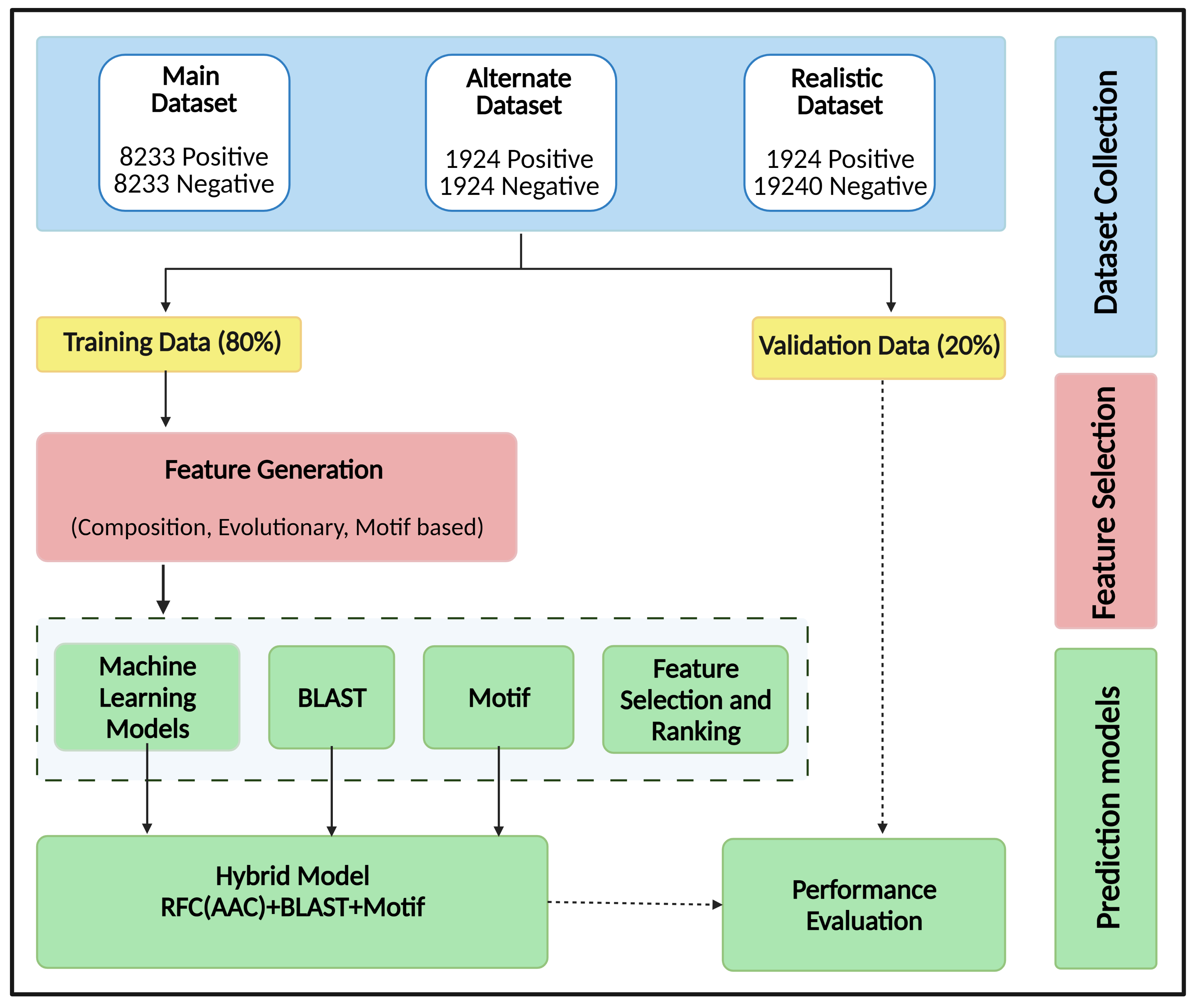
We have collected the positive (toxic) and negative (non-toxic) data from UniProt. We have created three datasets based on the number of toxic and non-toxic protein sequences, as described below:
(a) Main Dataset: 8233 toxic and 8233 non-toxic protein sequences
(b) Alternate Dataset: 1924 toxic and 1924 non-toxic protein sequences
(c) Realistic Dataset: 1924 toxic and ten times negative dataset, i.e., 19240 non-toxic protein sequences
The data was divided into training and validation dataset in the ratio of 80:20. We have extracted the Composition, Evolutionary and Motif based features on these dataset and build the machine learning models. We have also applied BLAST tool for identifying the toxic proteins. In addition to this, we have also searched for motifs in the dataset using MERCI software.
Further, we have also implemented a hybrid approach to enhance the prediction of the model. For this, the following three techniques have been integrated: (i) similarity-based approach using BLAST, (ii) motif-based approach using MERCI and (iii) Machine learning-based technique. We have implemented the best performing models for predicting toxic and non-toxic protein at the web interface.