Help
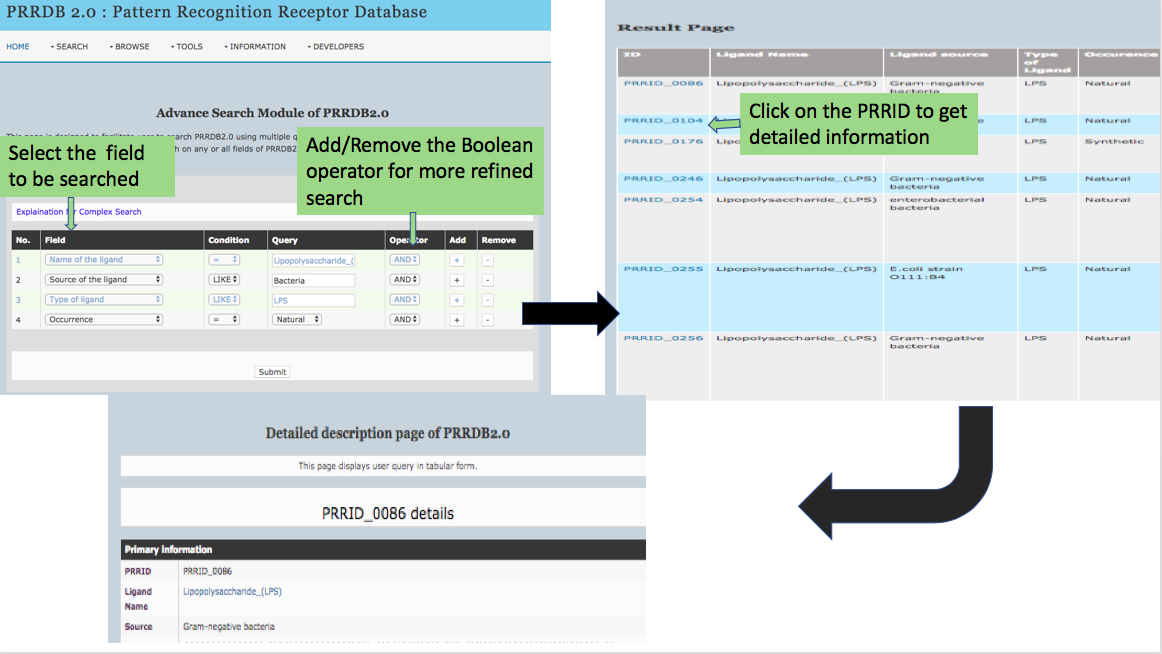
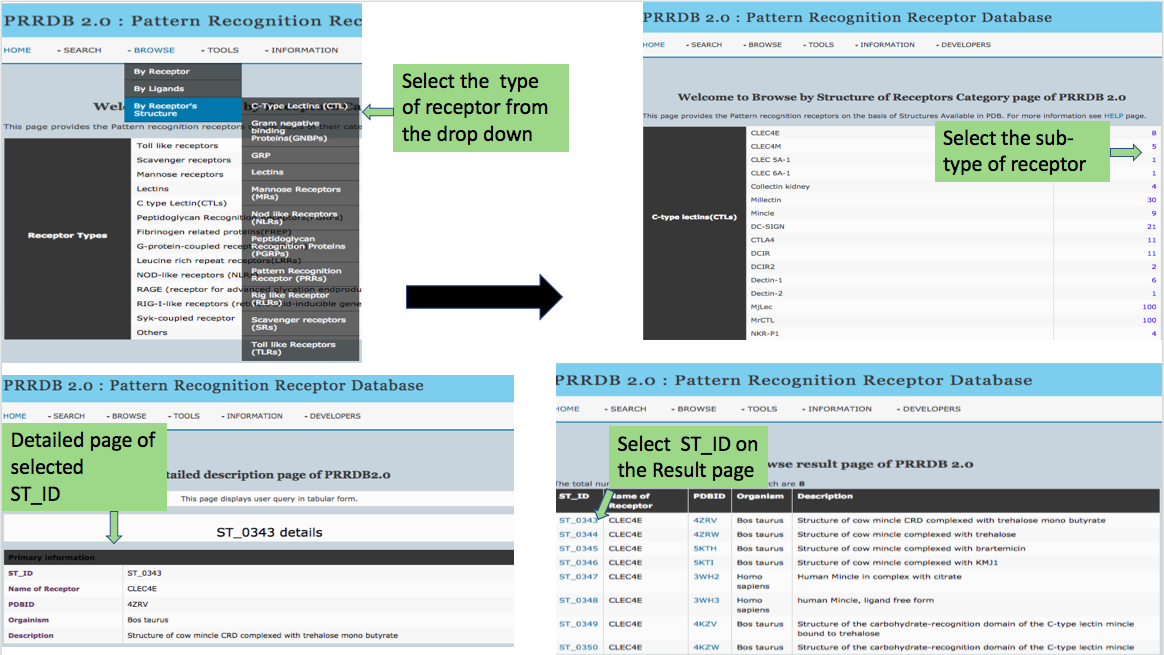
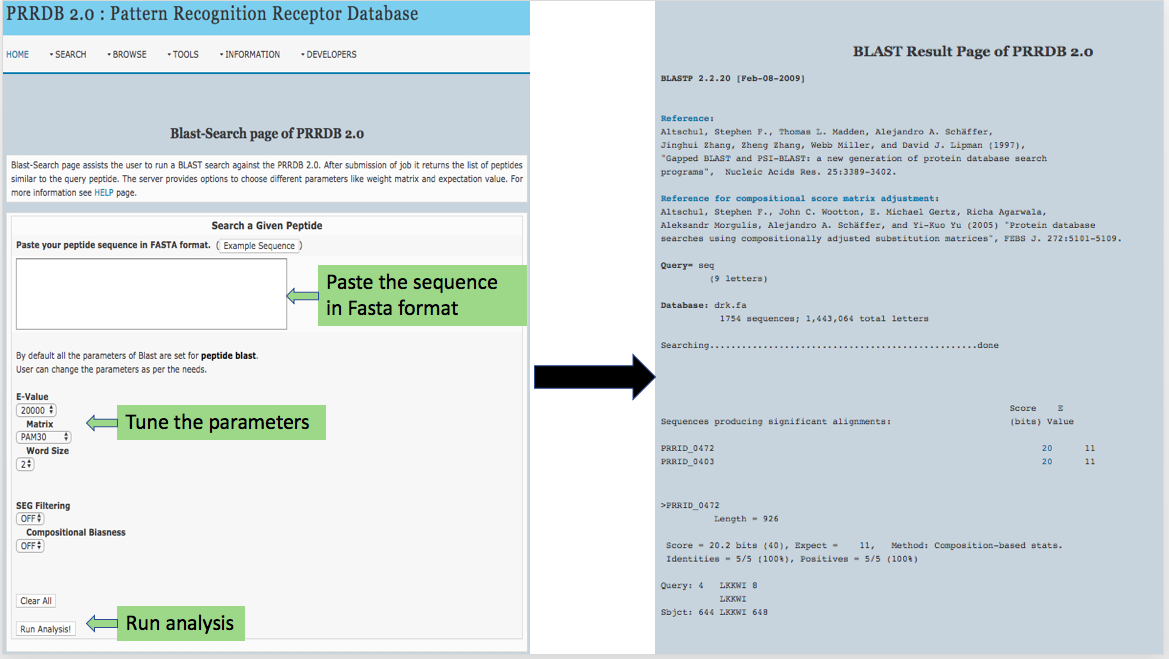
This page provides help to the users of PRRDB 2.0 database on how to efficiently use this repository of pattern recognition receptor. For each module which is implemented in PRRDB 2.0, here we provide the figures and description on how to use that module. Four general modules are provided here.
Frequently Asked Questions (FAQs) |
|
Q1. What is PRRDB 2.0? Ans. PRRDB 2.0 is an updated version of PRRDB (released in 2008) which is a comprehensive repository of experimentally verified pattern recognition receptors and their ligands. Q2. Why PRRDB 2.0? Ans. It comprises more than 2000 entries from the literature available on Pubmed. It gives all the information related to pattern recognition receptors like its Ligand, source of ligand, function of ligand,occurence of ligand, receptor, receptor source, Function of receptor, assys used, etc. Q3. How to search into PRRDB 2.0? Ans. A user can search by Ligand name or search by Receptors name. Facility of simple search and advanced search is also available for user friendly experience. |