This webserver has been designed to help the scientific community in designing lytic phage based therapy against pathogenic bacterial infections. This server helps to predict bacterial host targets for query phages, potential lytic phages for query bacteria and interactions between query phage-bacteria pairs. This page provides help on different modules of server. Following are major modules in this server:
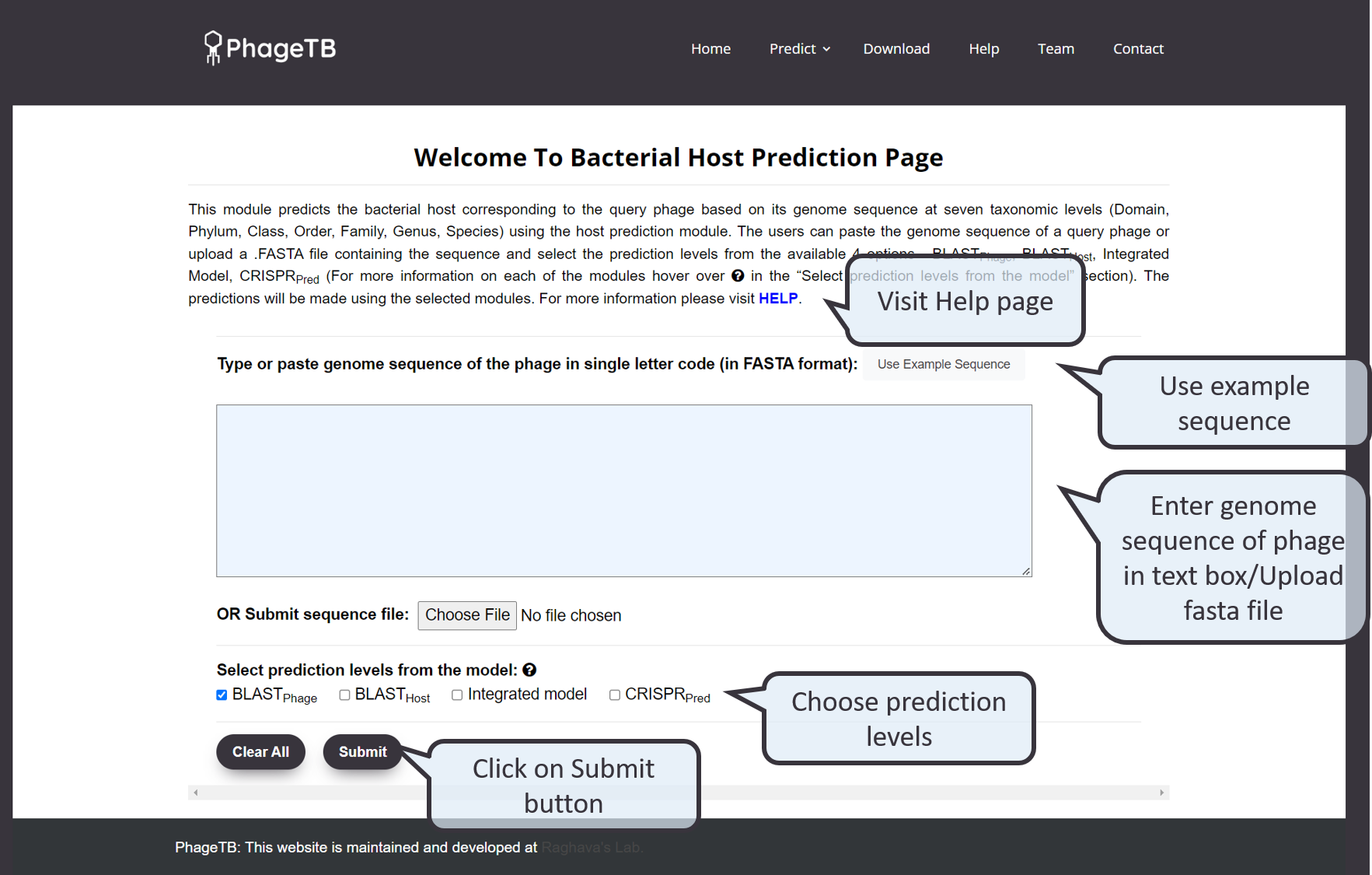
This module allows users to predict the bacterial hosts corresponding to the query phages. The user has to enter the genome sequence of the phage in the text box/upload a fasta file contiaining the geneome sequence. The users are to allowed to select one or more methods from the prediction stack of PhageTB they want to use for making the predictions using the check boxes provided. The server predicts the host bacteria from its reference database of bacterial hosts, enlisting it at various taxonomic levels

This module allows users to predict whether given phage and bacterial hosts are likely to interact with one another. The user has to enter the genome sequences of the phage and bacteria in the text boxes/upload a fasta files contiaining the geneome sequences. The users are to allowed to select one or more methods from the prediction stack of PhageTB they want to use for making the predictions using the check boxes provided and Enter the e-value for BLAST alignment for the predicted and query bacterial hosts, based on which the prediction of interaction will be made. The server predicts the target phage from its reference database of interactions, and also provides the reference bacterial host of the predicted phage with the measure of similarity with the query bacteria


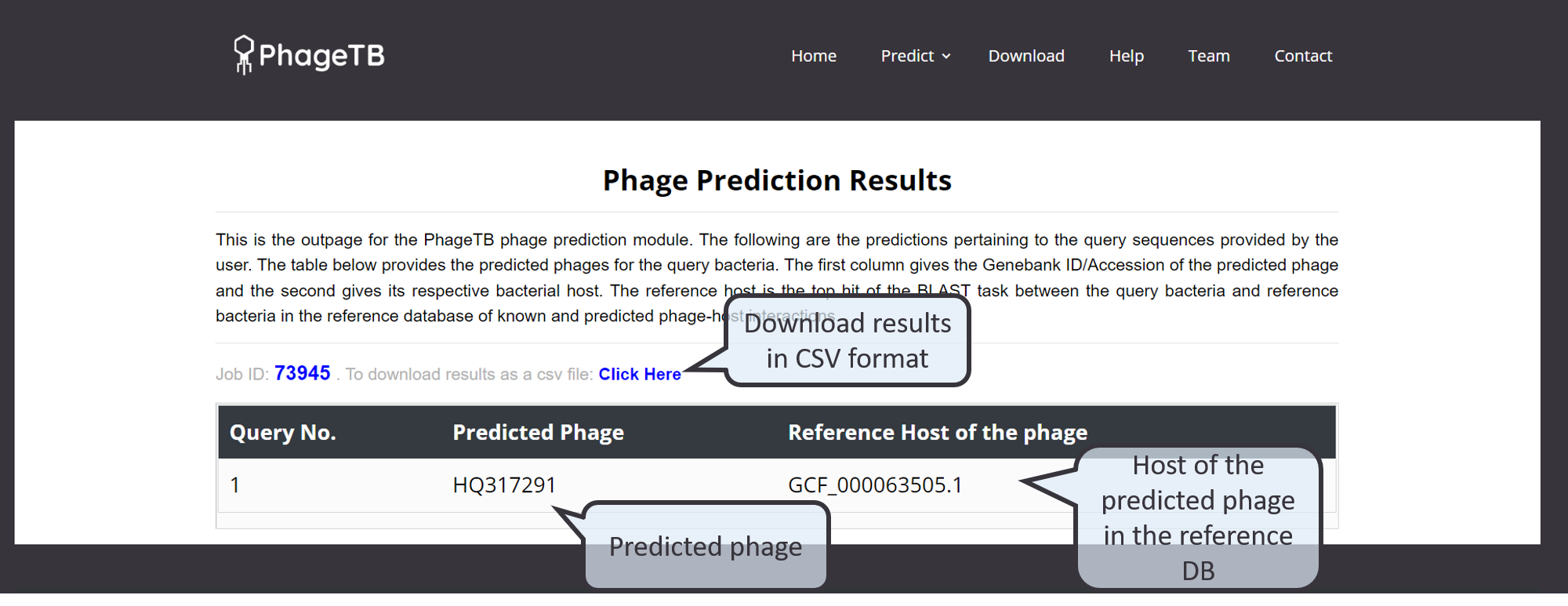
This module allow users to predict the target phage likely to infect query bacteria. The user has to enter the genome sequence of the bacteria in the text box/upload a fasta file contiaining the geneome sequence. The users are to allowed to select the evalue for BLAST alignment of query bacteria with reference bacterial hosts, based on which the top hit(s) are used for predicting the target phages. The server predicts the target phage from its reference database of bacteriophages, and the top hit reference hosts

PhageTB: This website is maintained and developed at Raghava's Lab.