This page provides help to the users of CPPsite 3.0 on how to effectively use this database. For each module which is implemented in CPPsite 3.0, here we provide the figures and description on how to use that module. A user can click on the links given below to get help of the respective modules.
The Enquire module allows you to search and retrieve specific information.
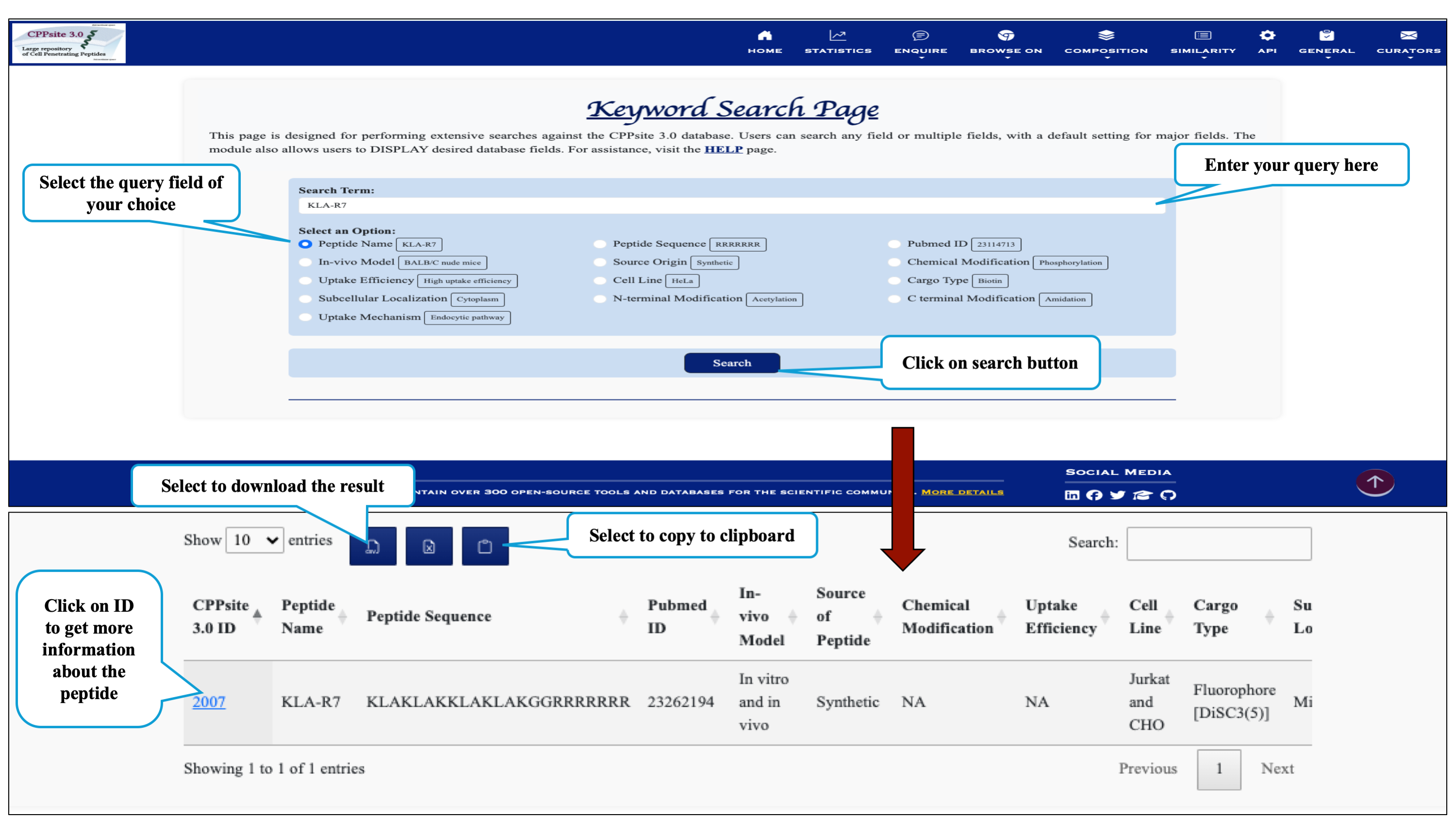
This module displays the simple search page of CPPsite 3.0 by selecting specific keywords. Clicking on the required field will allow the user to browse the specific information in the database.
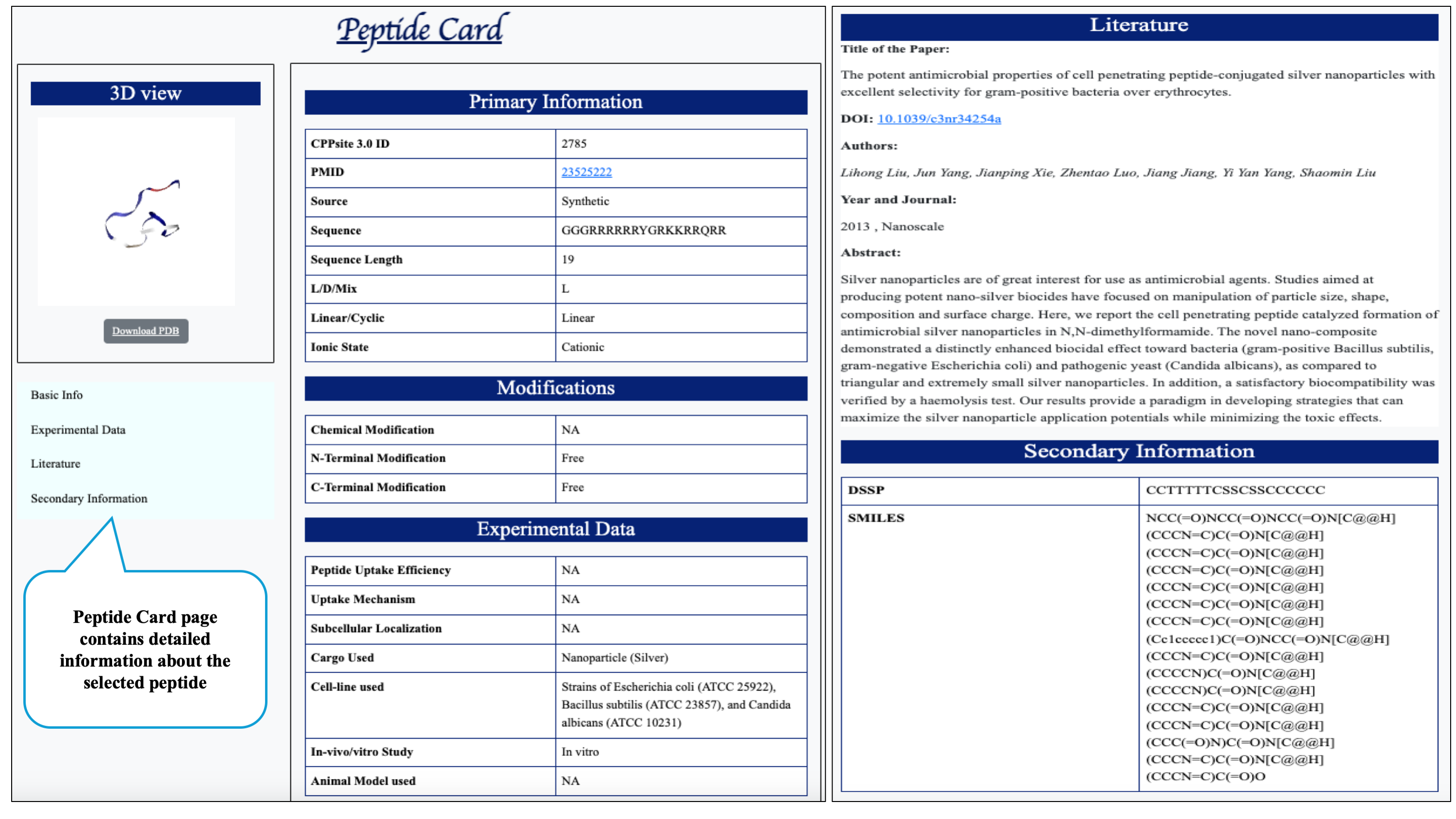
On clicking peptide ID, the peptide card page will display which contains detailed information about the peptide from its basic description, literature paper to its SMILE format.
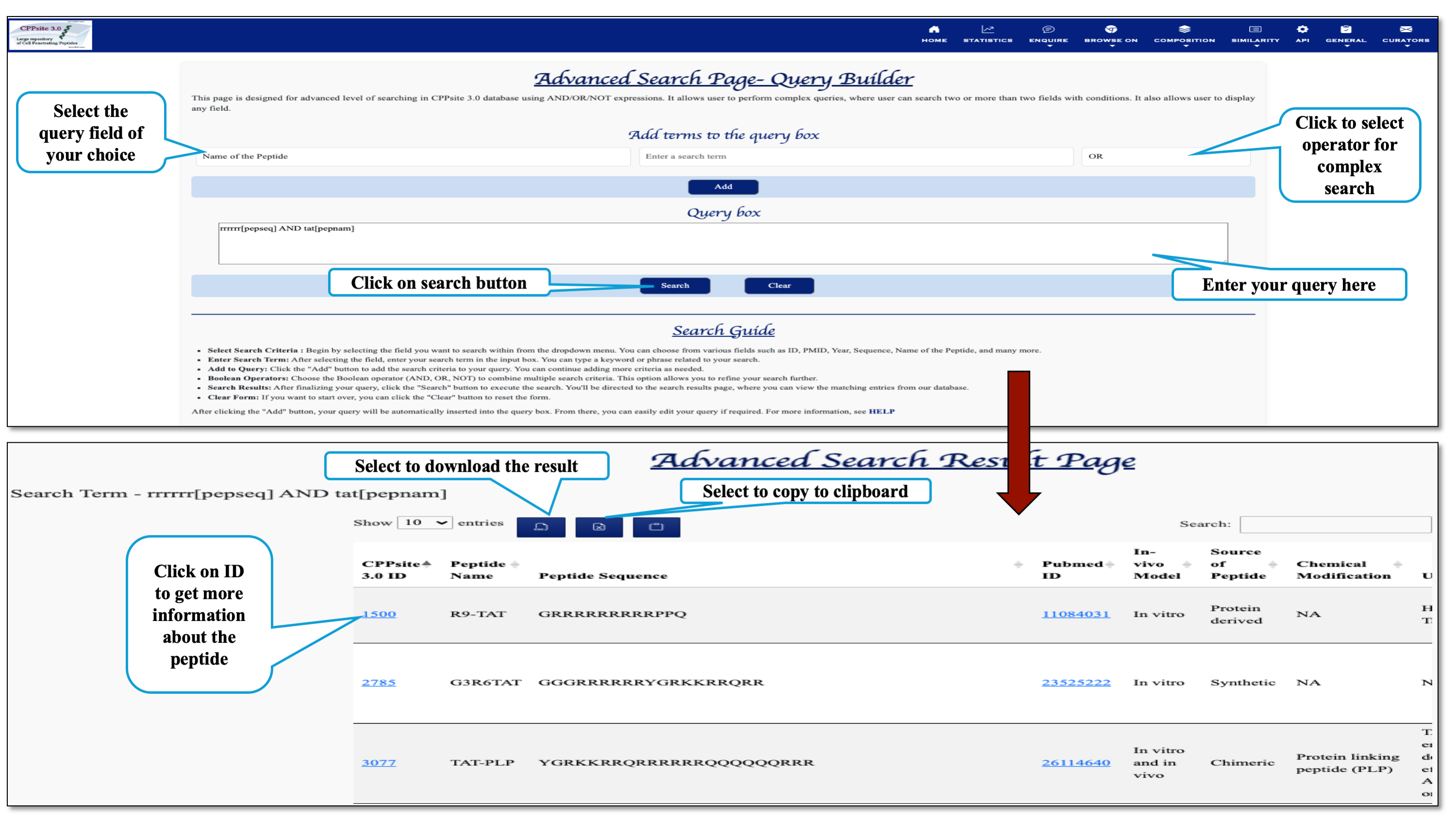
This page provides the advanced search facility in CPPsite 3.0 with the complex query generated by users using Boolean expression. User can browse the page by selecting two or more than two fileds.
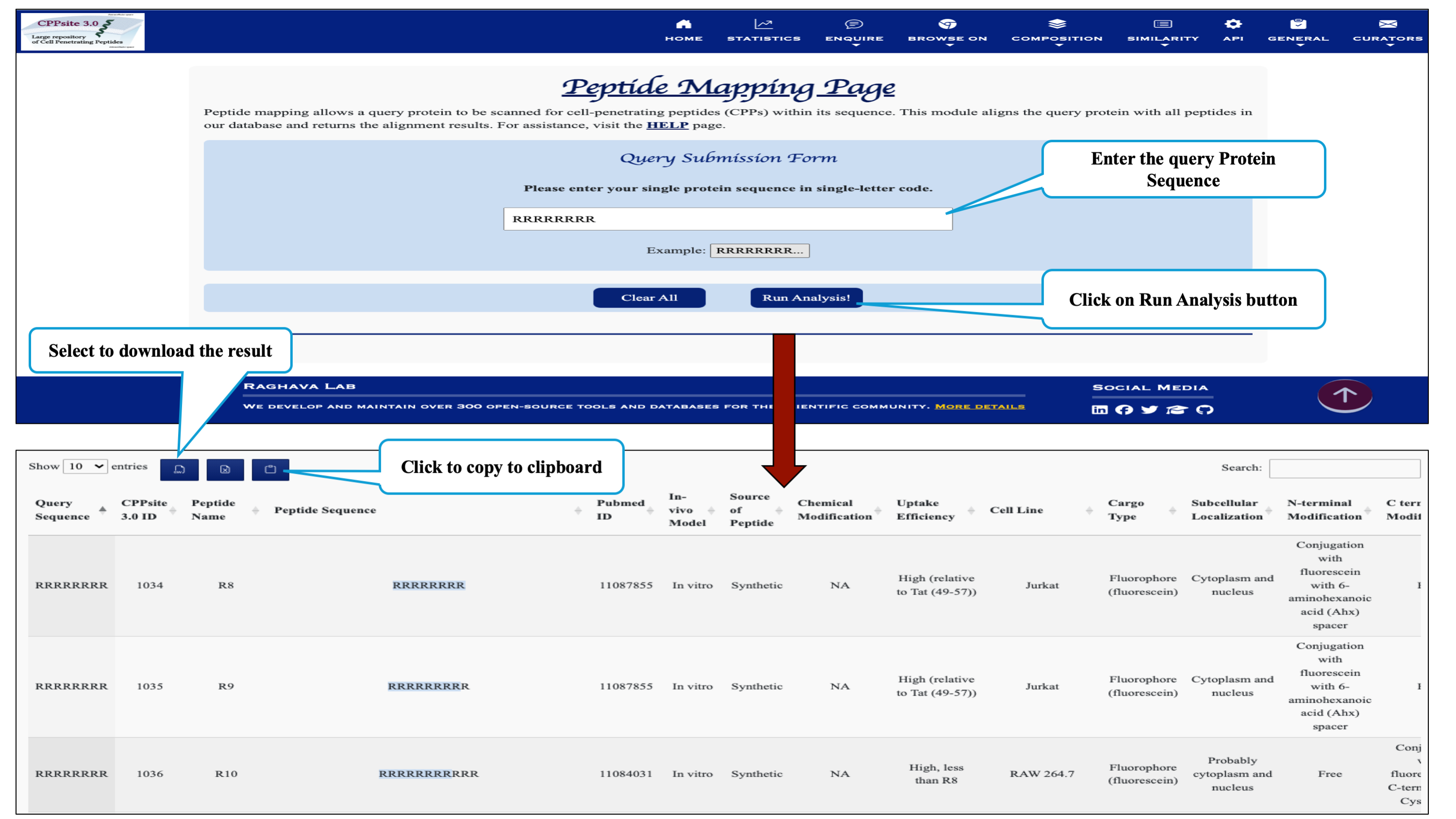
This page provides facility to search protein sequence having cell penetrating peptide segments and returns the alignment of the query sequence with all the peptide sequences present in the CPPsite 3.0
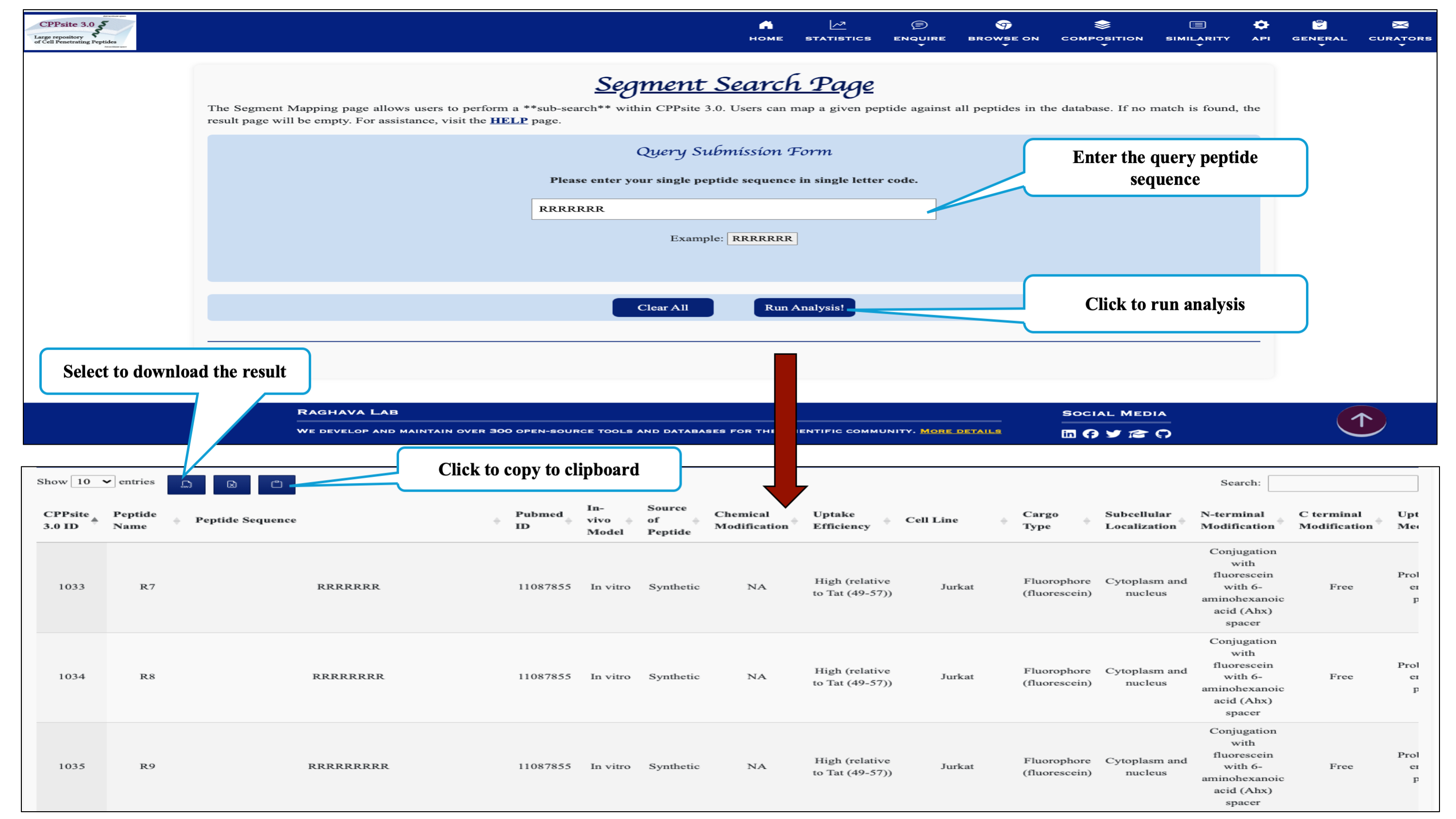
This page provides the facility of mapping the given query sequence with the cell penetrating peptide sequences present in the database CPPsite 3.0.
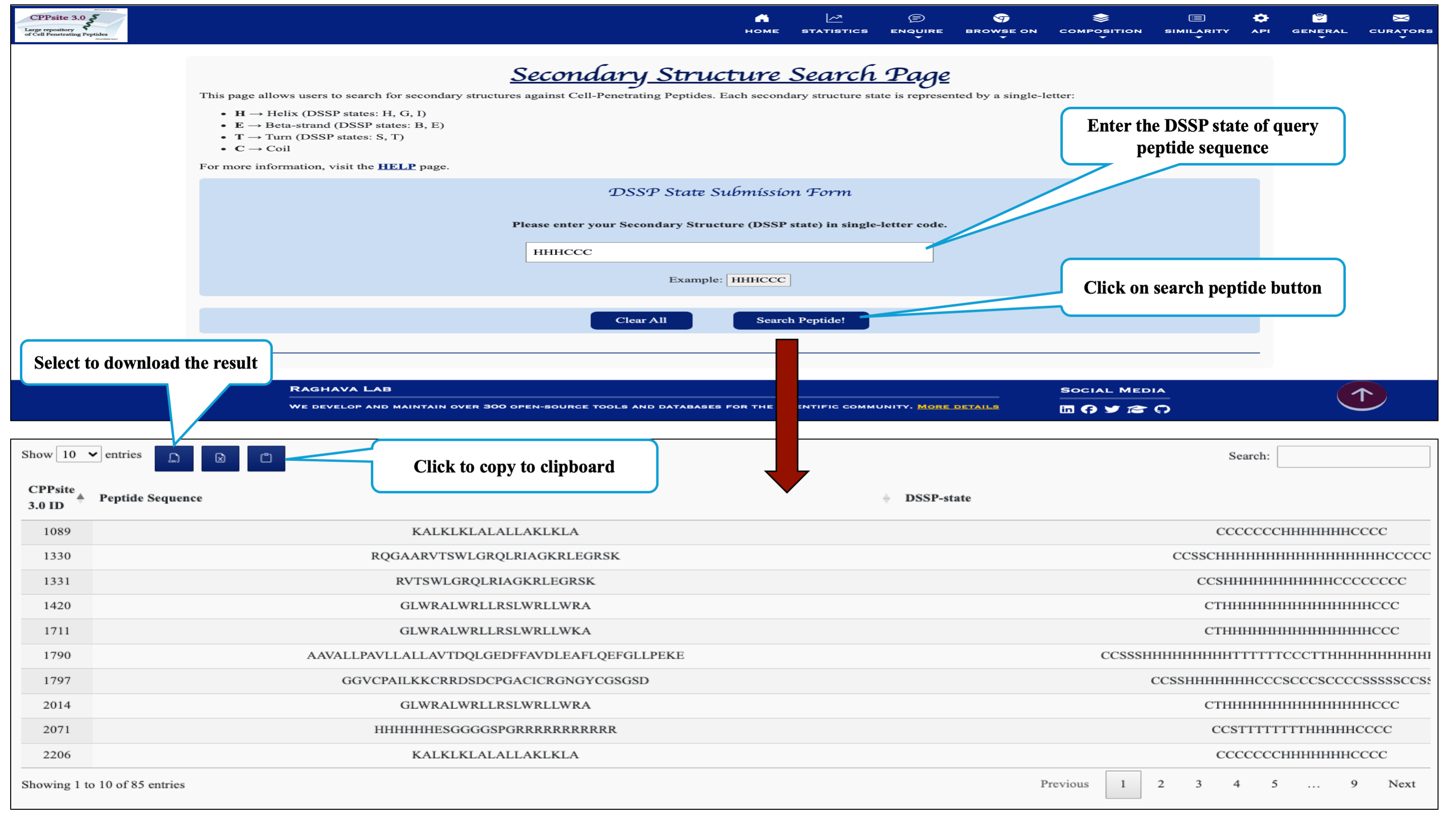
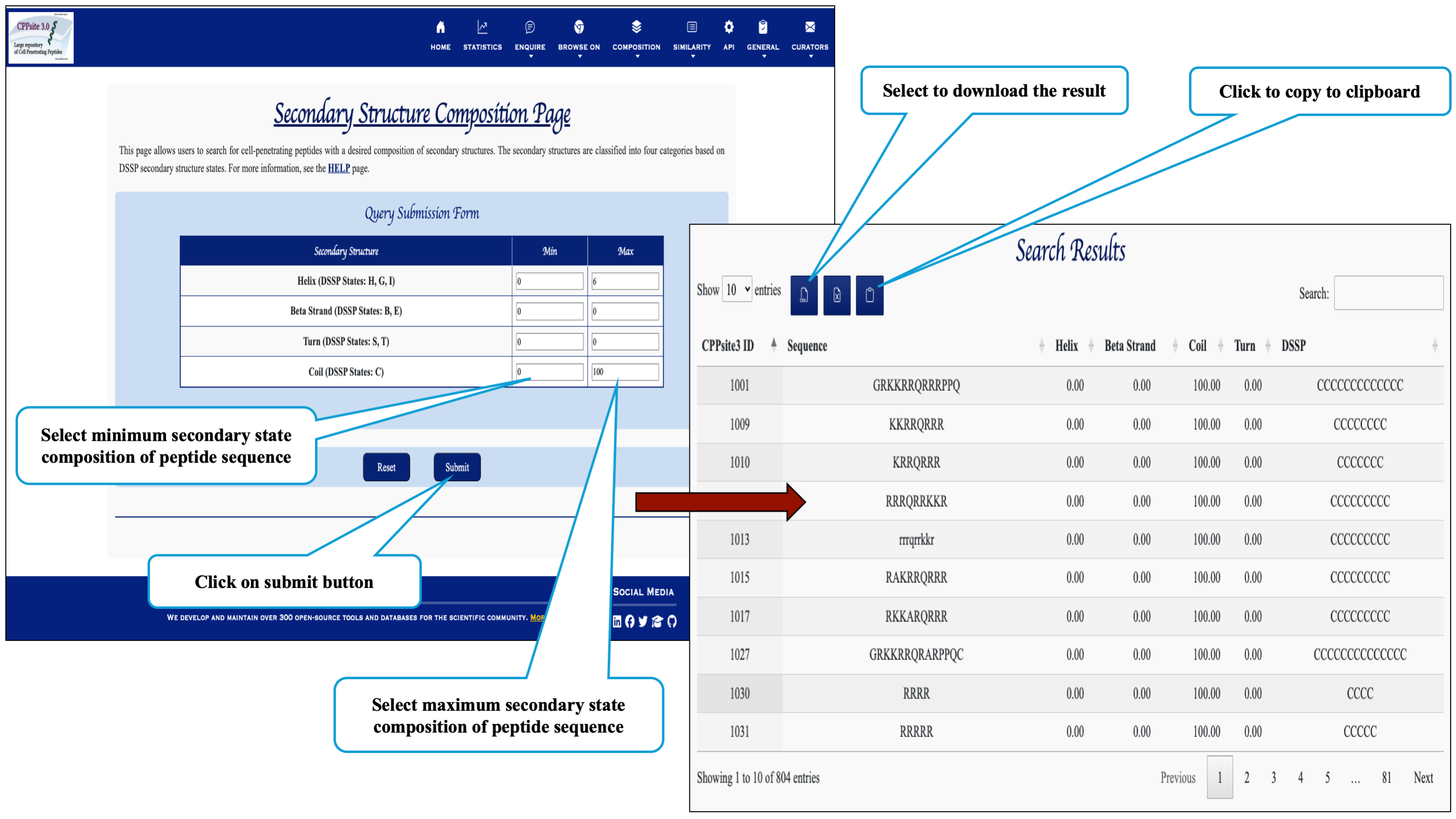
This page provides the faciltiy to search user's secondary structure against CPPs. User can submit its query sequence in DSSP state and search it in the database.
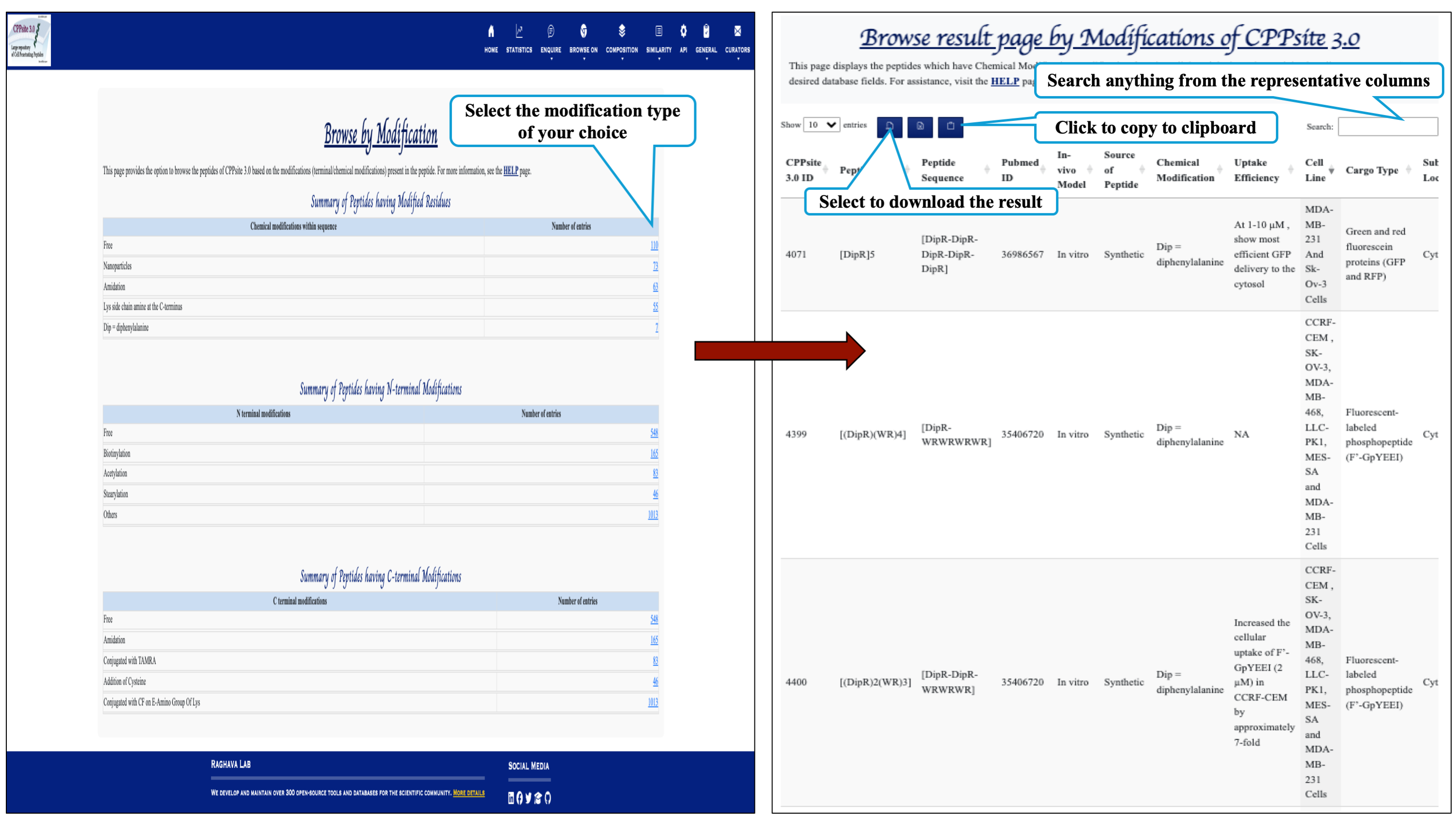
The Browse On module enables you to explore information based on modifications, catergory and peptide length.
This page provides the distribution of different modifications (Terminal modifications, Various chemical modifications) present in the peptides of CPPsite 3.0 database.
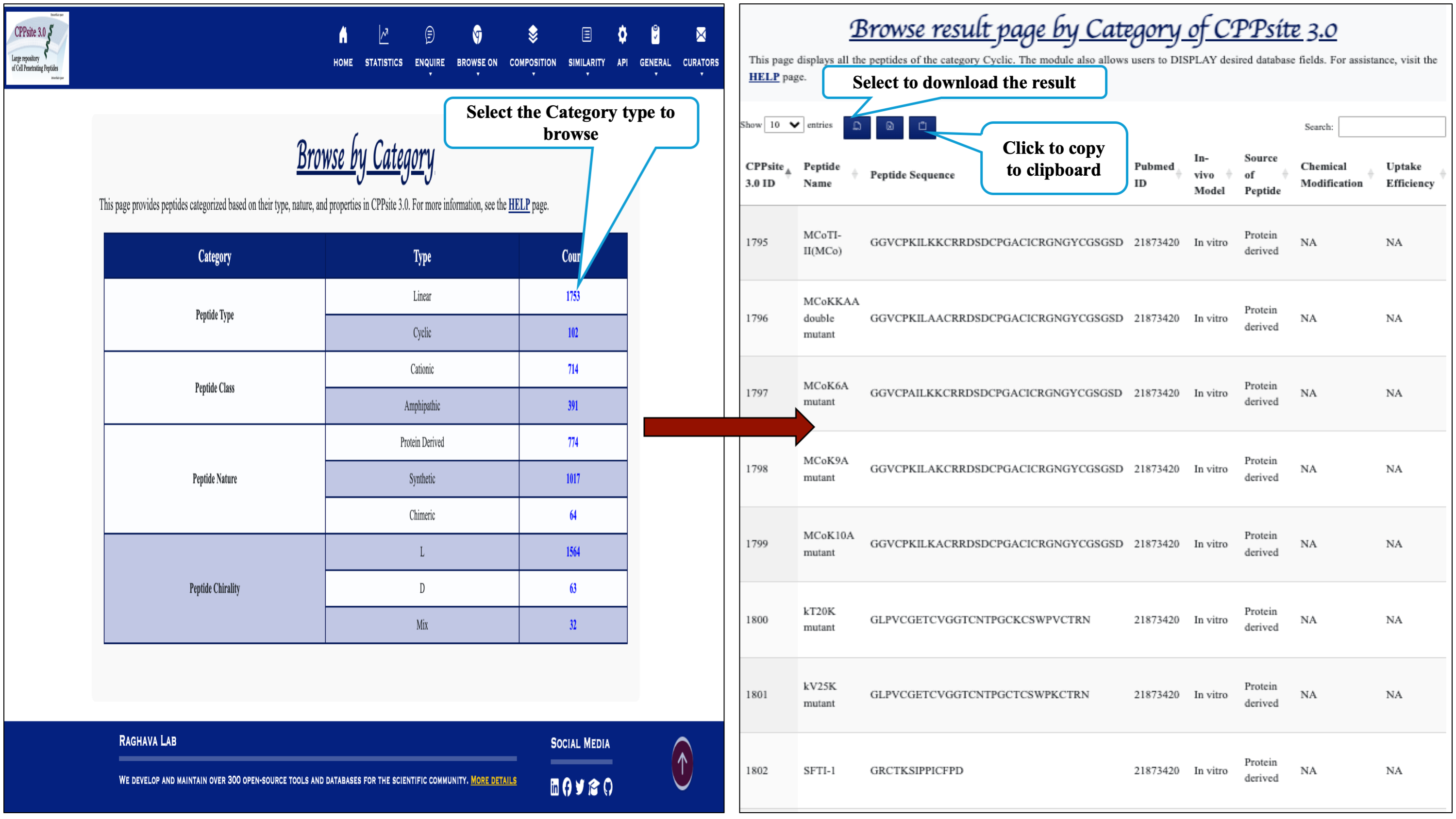
This module displays the category of peptides present in the database. Clicking on number gives the details about the type of peptide selected.
This module displays the peptides of different length, present in the database. Clicking on number gives the details about the type of peptide selected.

The Composition module enables user to explore amino acid compositions and their functional properties.
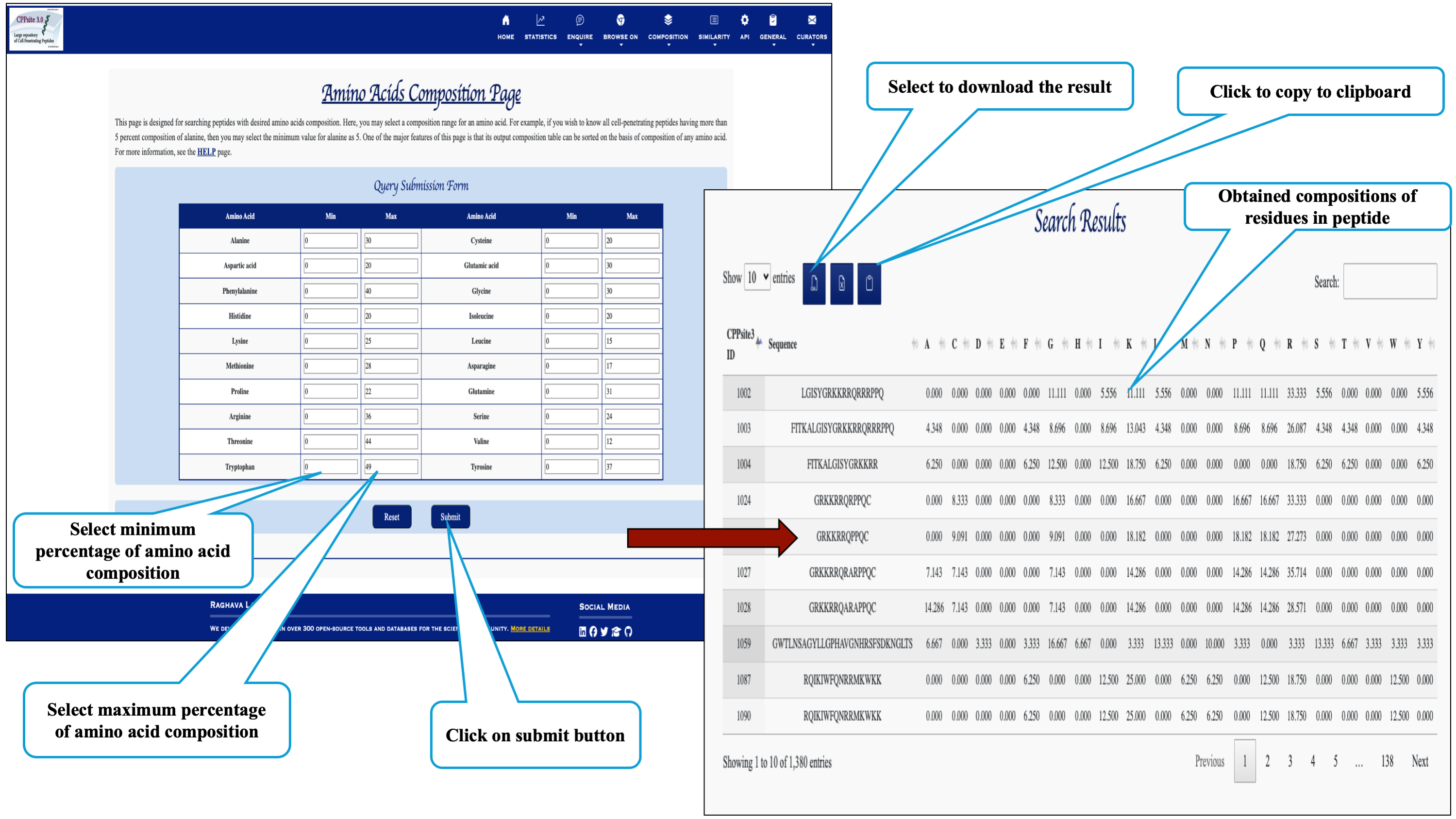
Here the user can browse the peptides with desired amino acid composition.
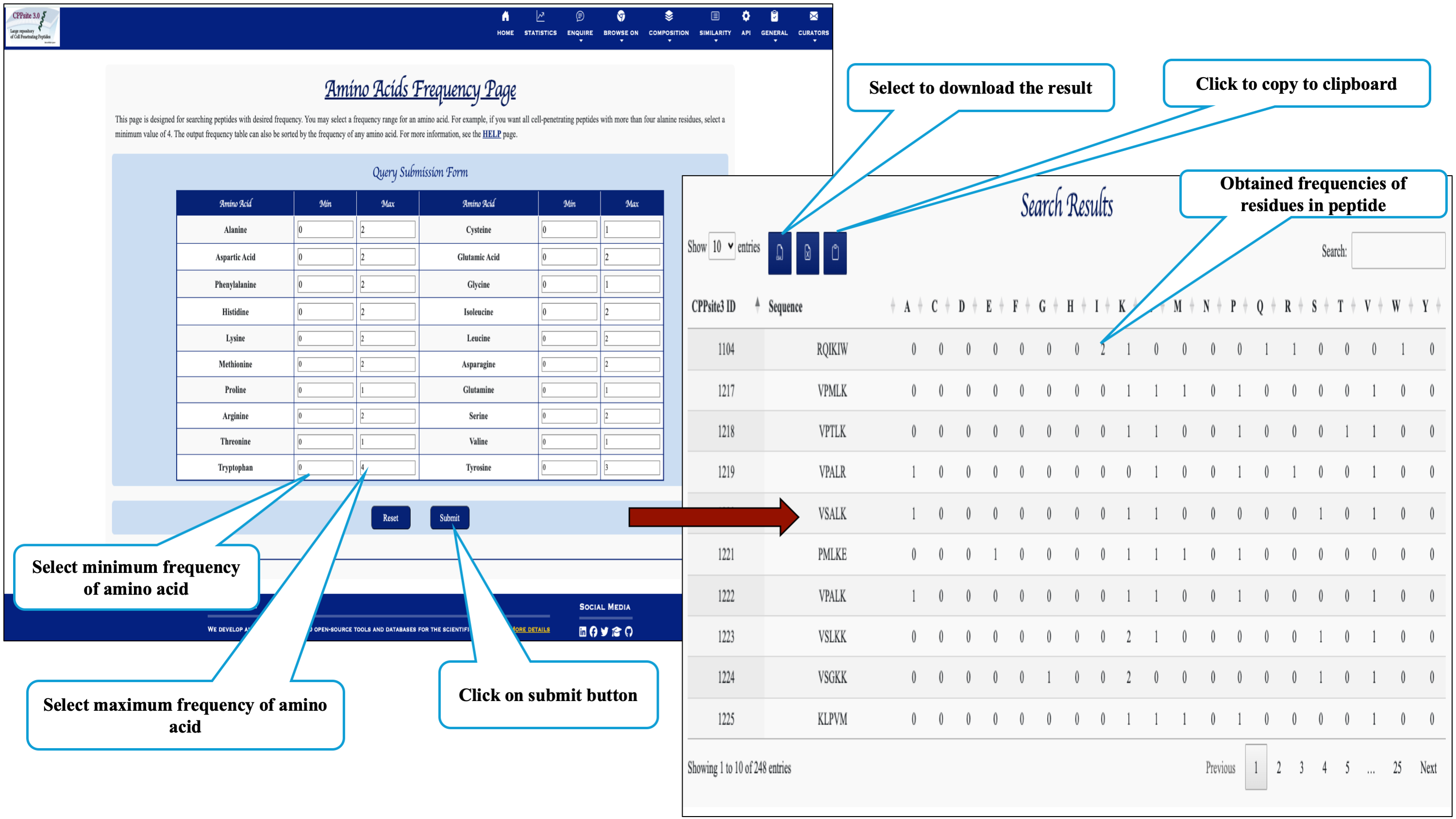
Here the user can browse the peptides based on the frequency of amino acid.
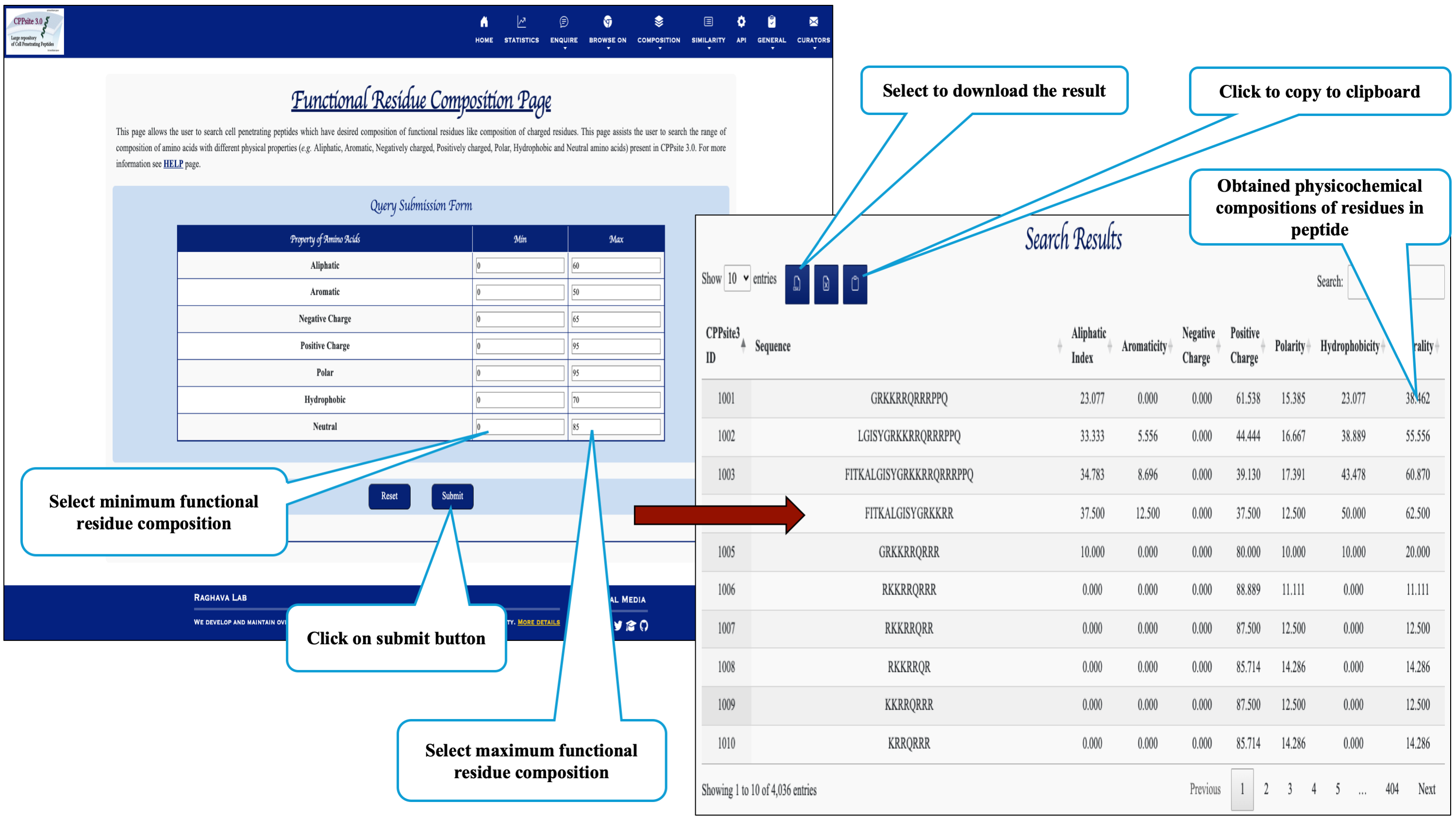
Here the user can browse the peptides with desired physiochemical composition.
Here the user can browse the peptides based on the frequency of physiochemical properties.
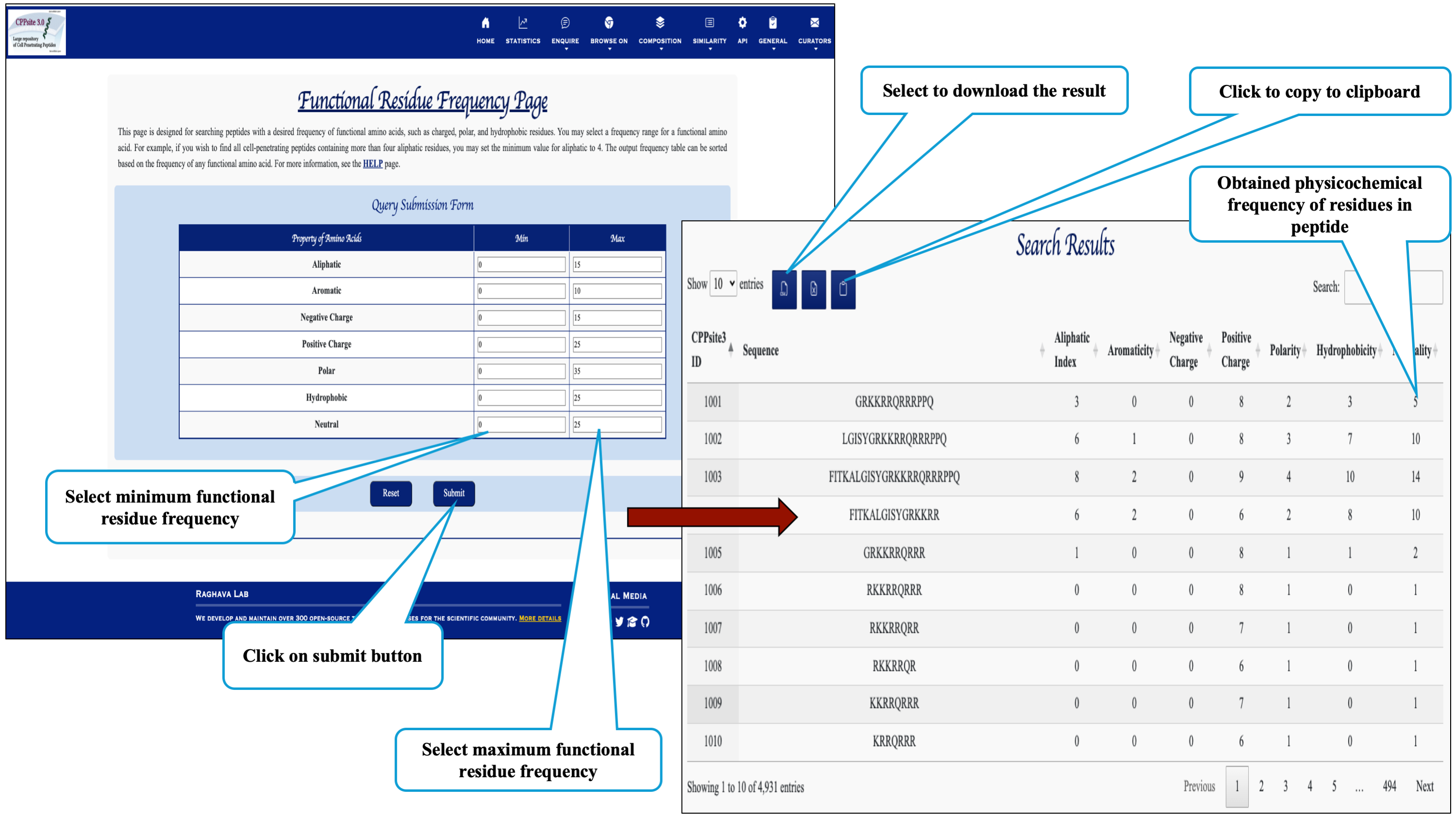
This page assits the user to search cell penetrating peptides based on their secondary structure composition.
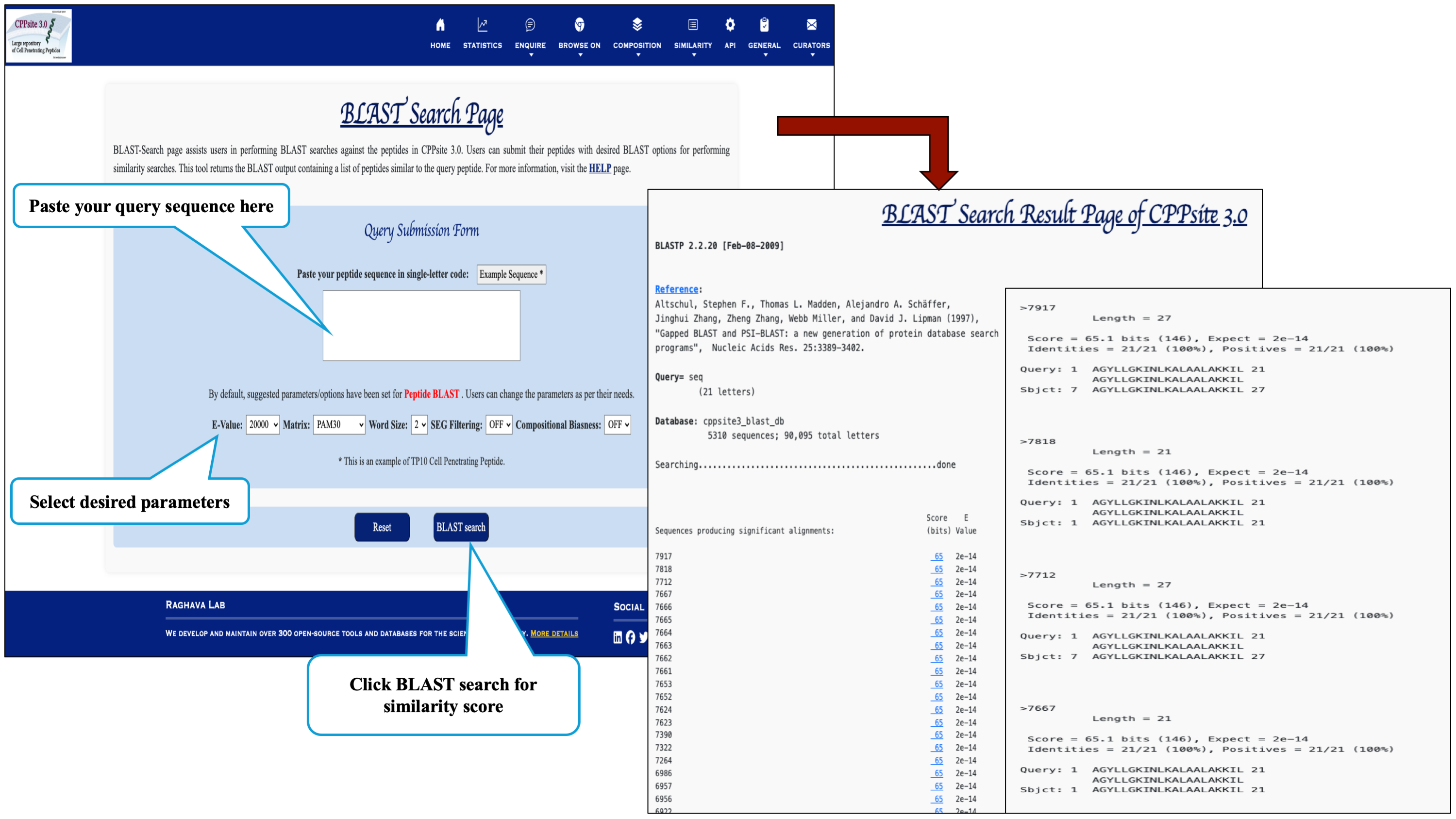
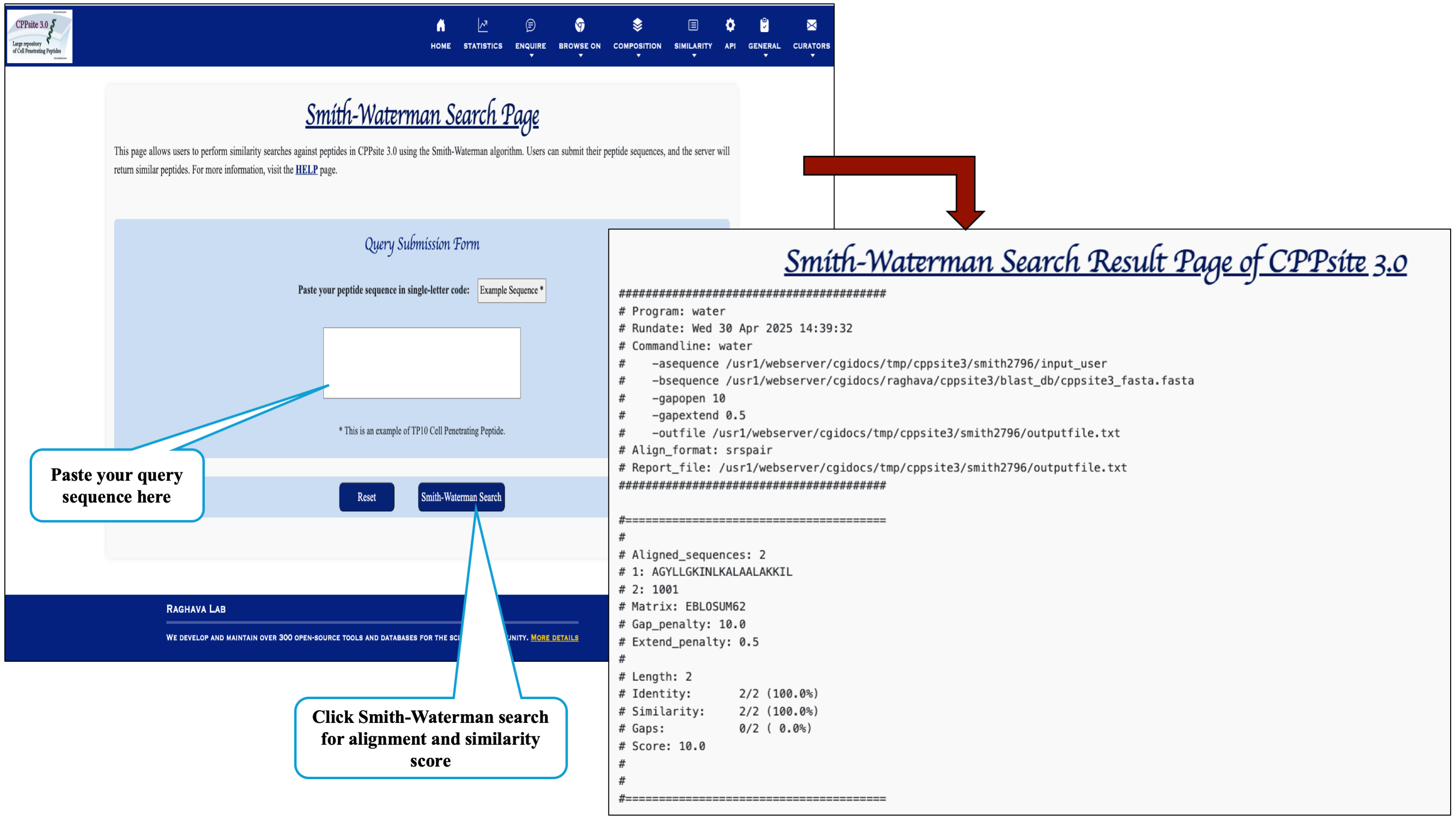
The Similaroty search tools helps in identifying similar peptid sequences to that of cell penetrating sequences available in the database.
Blast-Search page assists users in performing BLAST of the query peptide which will returns the BLAST output containing list of peptides similar to the query peptide.
Users can run a Smith-Waterman search query against the CPPsite 3.0 database. After submission of job it returns the list of similar peptides.
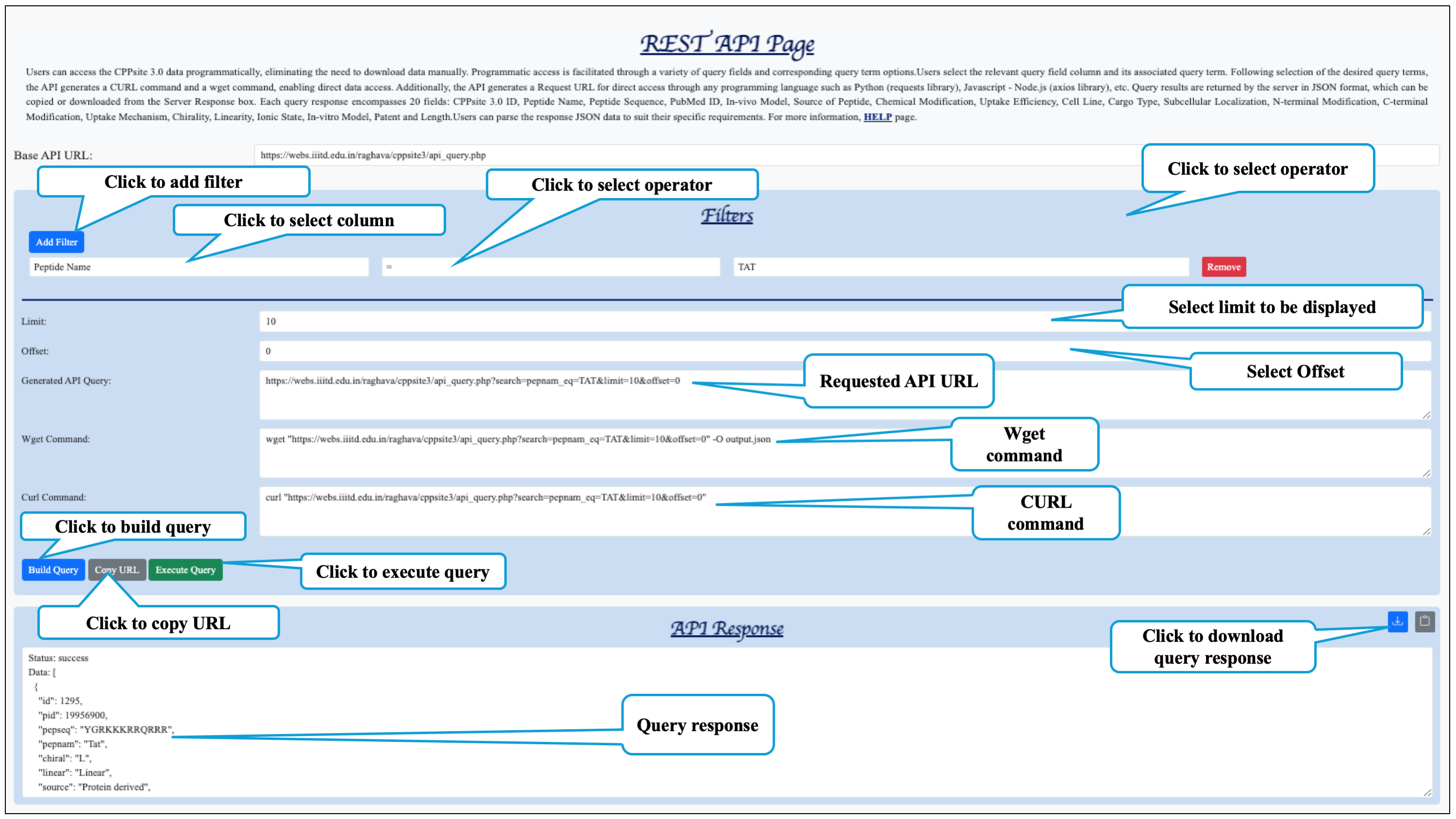
This section describes how this website's data can be accessed with programs. A variety of data
available on CPPsite 3.0 is accessible using simple URLs (REST) that can be used in
programs.
The CPPsite 3.0 REST API returns the response in JSON (JavaScript Object
Notation) format. Users can parse the JSON format to suit their requirements.