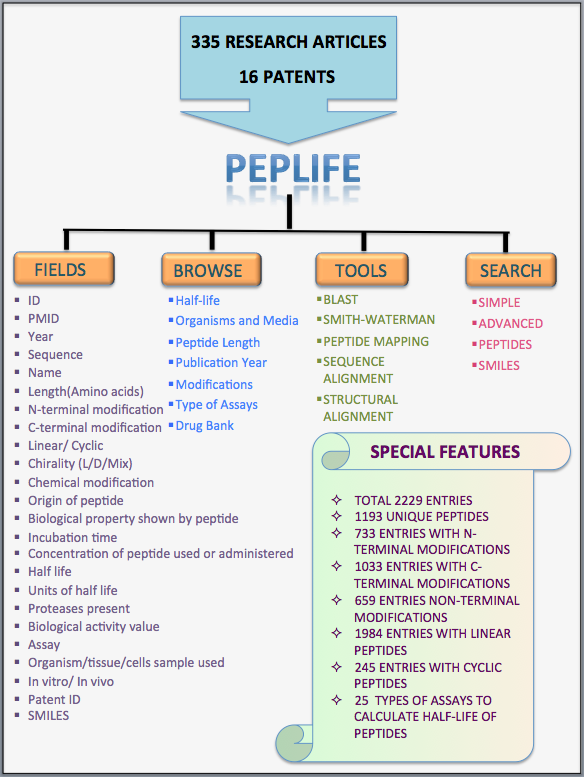
Brief description of all the fields in PEPlife
| Name of the Field | Description |
|---|
| Name of peptide | This field gives the name of the peptide as mentioned in the article. |
| Article PMID | This provides the PMID of the article from which the entry is made. |
| Publication year | It gives the year in which the article was published. |
| Peptide sequence | This field gives the sequence of the peptide. |
| N-ter modification | It gives information regarding the N-terminus modification incorporated in the peptide. E.g. Acetylation |
| C-ter modification | It gives information regarding the C-terminus modification incorporated in the peptide. E.g. Amidation |
| Cyclic/Linear | This field describes whether the peptide is linear or cyclic. |
| Chirality | This fields provides the information whether the amino acids of the peptide have D conformation or L conformation or are Mix i.e. composed of both L and D amino acids. |
| Modification | This field gives the information about the type of chemical modification or non-natural amino acid incorporated in the peptide e.g. PEGylation |
| Origin of peptide | It describes from where the peptide originated e.g. Modified form of Vasopressin |
| Nature of peptide | It describes the biological property possessed by the peptide. E.g. Antimicrobial |
| Biological activity | It gives the value of the biological activity possessed by the peptide. E.g.IC50=0.006 µg/ml against IIIB virus. |
| Half-life of peptide | This field provides the half-life of the peptide. |
| Assay type | This field gives name of the assay used to check the half-life of the peptide. |
| Test sample | This field gives the details of the sample on which half-life was experimentallt validated. Eg Rat plasma. |
| in-vivo/in-vitro | This fields provides the information whether the experiment for determination of half-life was carried out in vitro or in silico. |
FAQs (Frequently asked questions)
Q1. What is PEPlife?
Ans. PEPlife is a manually curated repository of experimentally verified half-life of peptides. In this database, peptides have been collected from both published articles as well as from patents.Each entry provides comprehensive information about a peptide that includes its sequence, chemical modifications incorporated, biological activity possessed, its half-life, the assay used to determine its half-life etc.
Q2. Why is half-life of a peptide important?
Ans. Half-life of a peptide affects its bioavailability, bio-distribution and rate of clearance. It is a major factor in deciding the dosing regime of a peptide drug i.e. how much and how often it has to be administered. It is one of the major reasons why peptide drugs fail in clinical development.
Q3.Why was PEPlife created?
Ans. Peptide drugs are a growing area for therapeutics but one of the major challenges in its progress is the short life of peptides. Therefore, the main objective of creating PEPlife was to compile all the scattered information in literature in order to understand the properties that are responsible in determination of half-life of a peptide.
Q4. Does this database have the collection of all peptides’ half-life available in literature?
Ans. This database is the result of first round of curation and literature is being constantly searched with the objective of providing an exhaustive repository.
Q5. What is unique about PEPlife?
Ans. PEPlife is a unique and first database, which provides comprehensive information about half-life of peptides that are pharmaceutically important. It has many tools to facilitate users in analyzing different properties of peptides. PEPlife also provides 3D structures of peptides.
Q6. Why search PEPlife?
Ans. PEPlife provides all information of experimentally validated half-lives of peptides, details of the sample on which half-life was tested, route of administration and assay used to determine its half-life is mentioned as well. It gives details about the origin and nature of the peptides, its chirality, amino acid sequence, N- and C- terminal modifications.
Q7. How do I process a text search in PEPlife?
Ans. User can search a peptide by name, peptide sequence, assay type, test-sample, year and PMID etc.
Q8. Is this database useful if users have their own query sequences?
Ans. We appreciate user input. The easiest way for users is to submit their experimentally validated half-life of a peptide sequence on the submission page available on the database, which will be verified by our group. In particular, we encourage you to include complete information. This will minimize potential errors during data registration.
Q9. To whom can I report a discrepancy?
Ans. Please refer to the "Contact Us" page.