Framework of the study
EIPpred a quick and efficient activity prediction tool of inhibitory peptides in terms of minimum inhibitory concentration (MIC) values against E. coli., relying on a supervised machine-learning approach using sequence information. Twelve different regression models are implemented to predict the MIC value along with applied three different feature types to investigate the effect of features on the models. To remove irrelevant features, a standard method, mRMR, is performed for feature selection, which selects the top 1000 features that greatly contribute to the model’s performance. A robust k-fold cross-validation strategy was employed to analyse the regression models. Finally, the best model i.e., Random forest-based regressor built on 1000 selected features has been developed to predict the activity of the peptides. |
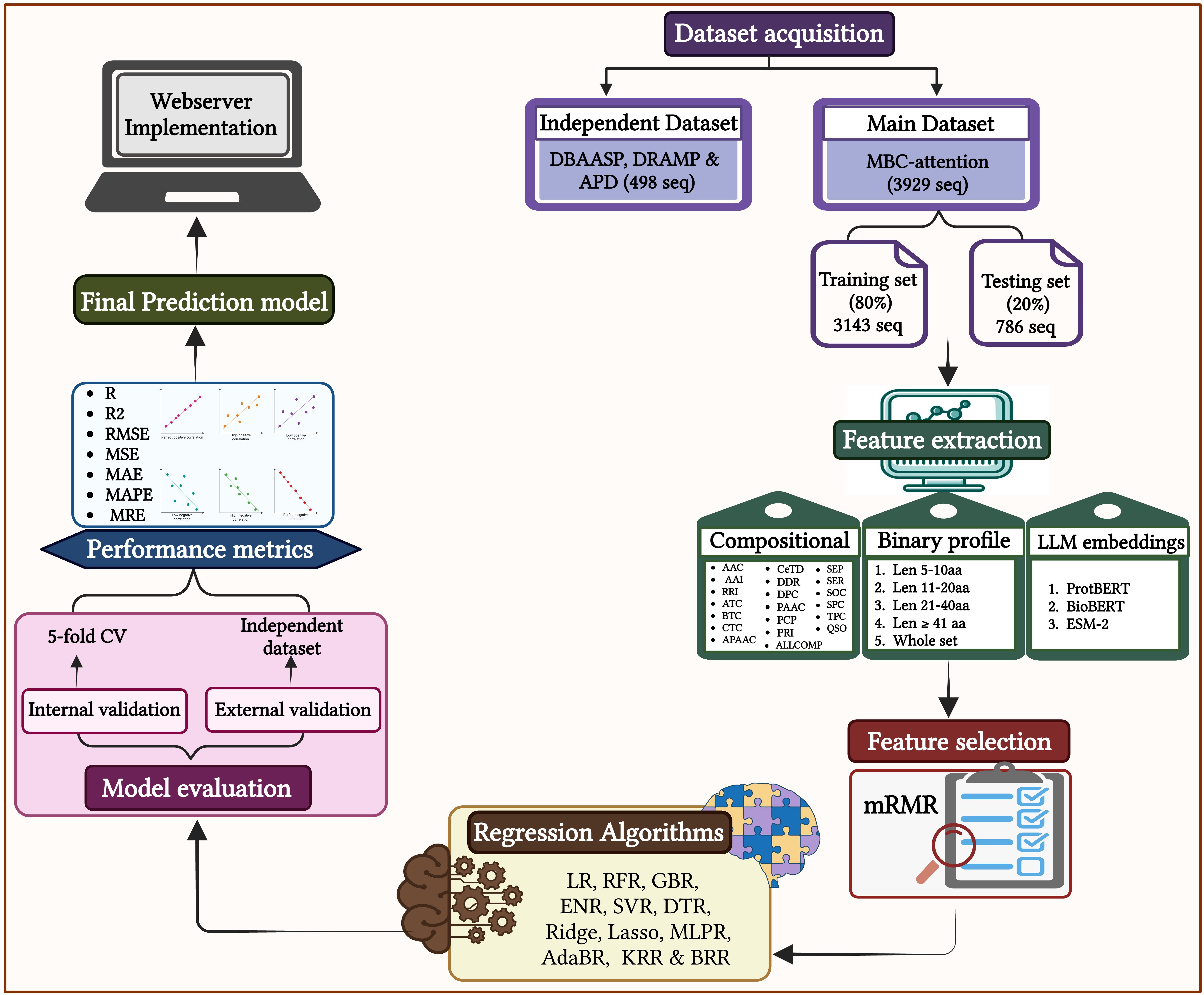
Datasets
Main Dataset
The training set comprised 80%, i.e., 3143 sequences of the total data, while the validation set was formed by the remaining 20% data, i.e., 786 sequences. We first arranged all peptides as per their length and then transferred the first peptide into the validation, then the next four in the train, again the sixth in the validation and so on.
Independent Dataset
We extracted 498 unique experimentally validated peptides that have been curated from DBAASP, DRAMP and APD databases with a count of 127, 342 and 29 peptides, respectively. We ensured none of these sequences were identical to the Main dataset