Welcome to Help Page
This page provides help to the users of CPPsite 2.0 on how to effectively use this dataset. For each module which is implemented in CPPsite 2.0, here we provide the figures and description on how to use that module. A user can click on the links given below to get help of the respective modules.
Enquiry Module |
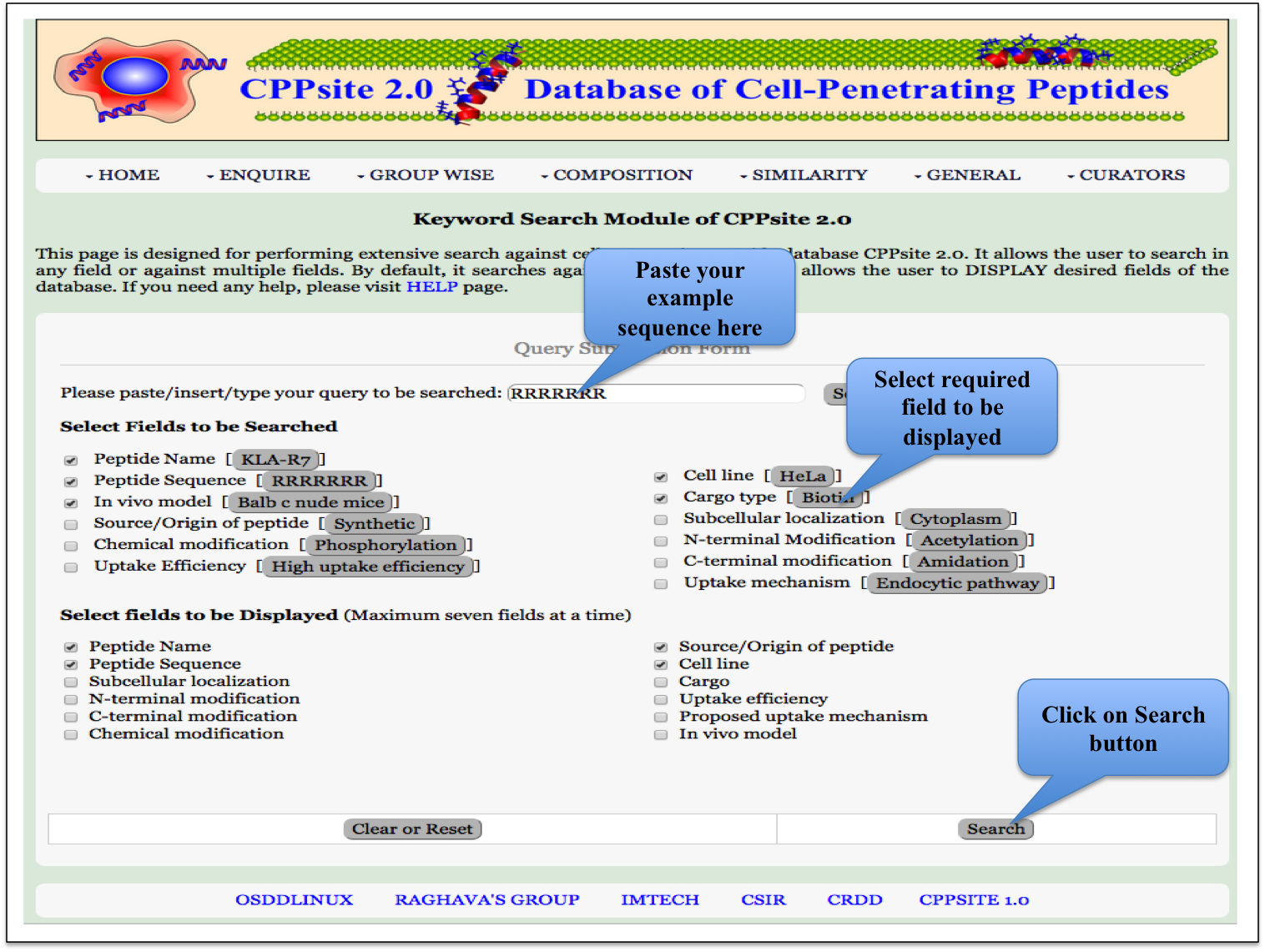
Keyword Search This module displays the simple search page of CPPsite 2.0. Clicking on the required field will allow the user to browse the specific information in the database. |
 |
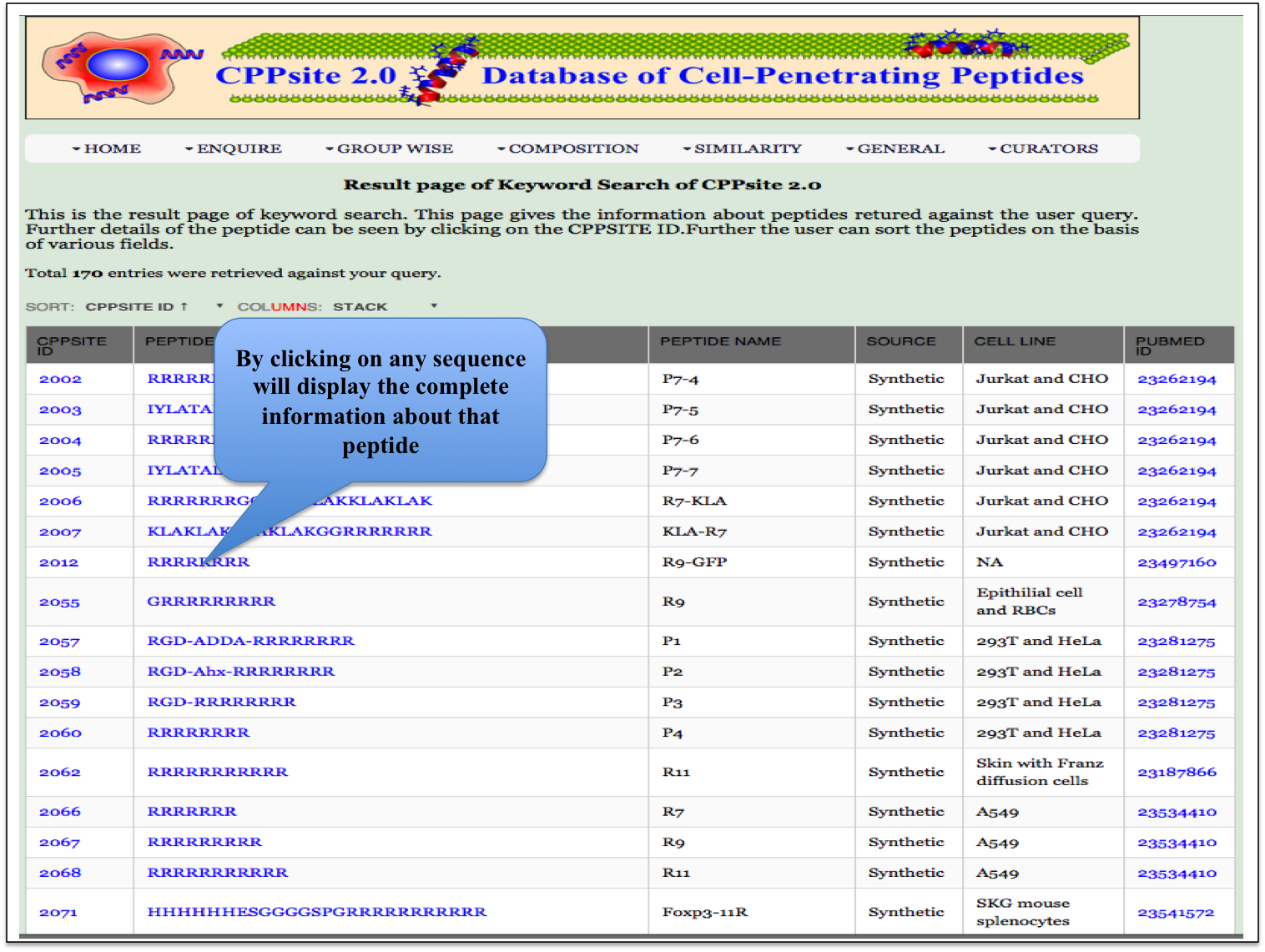
| Result |
 |
 |
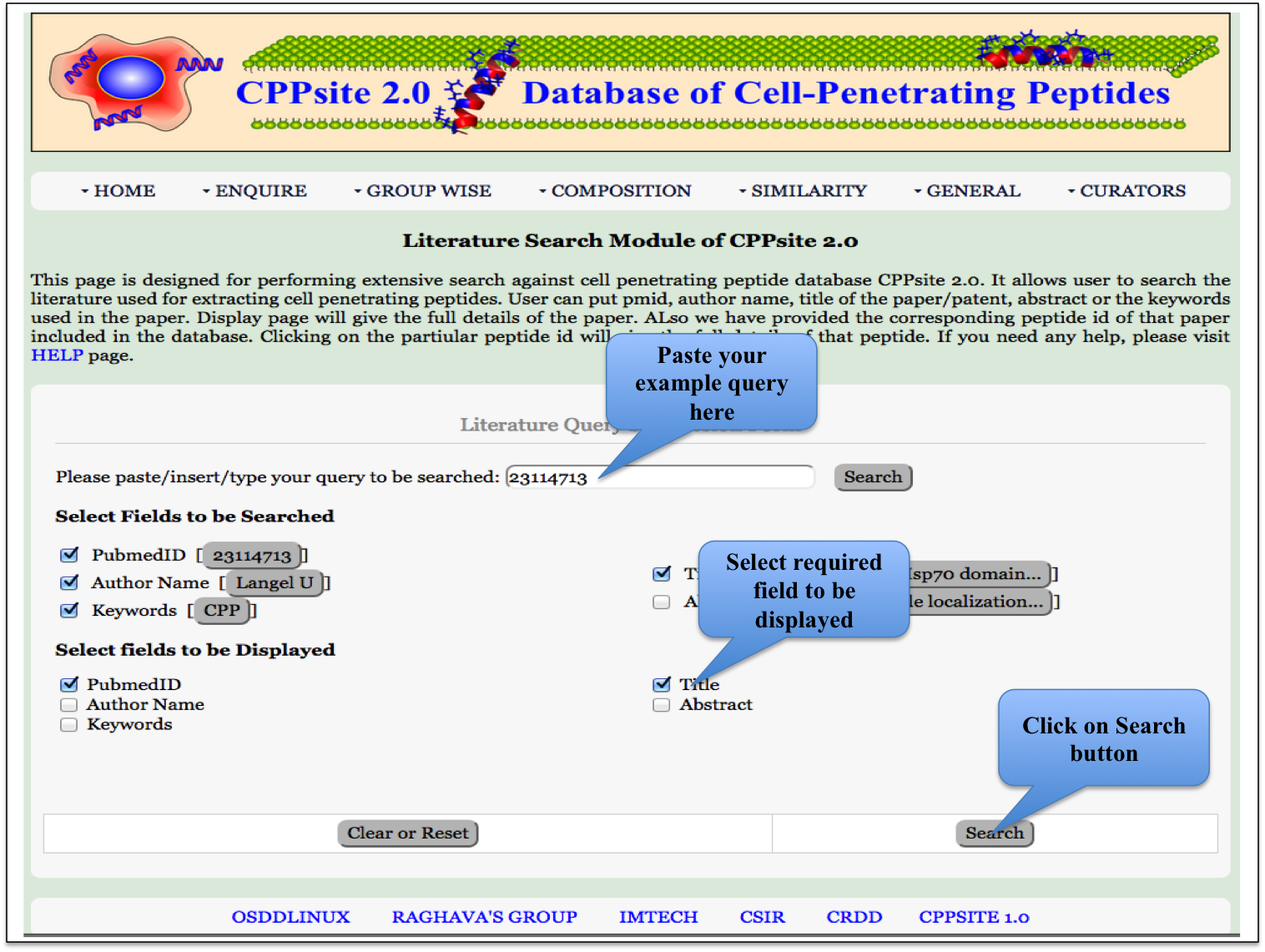
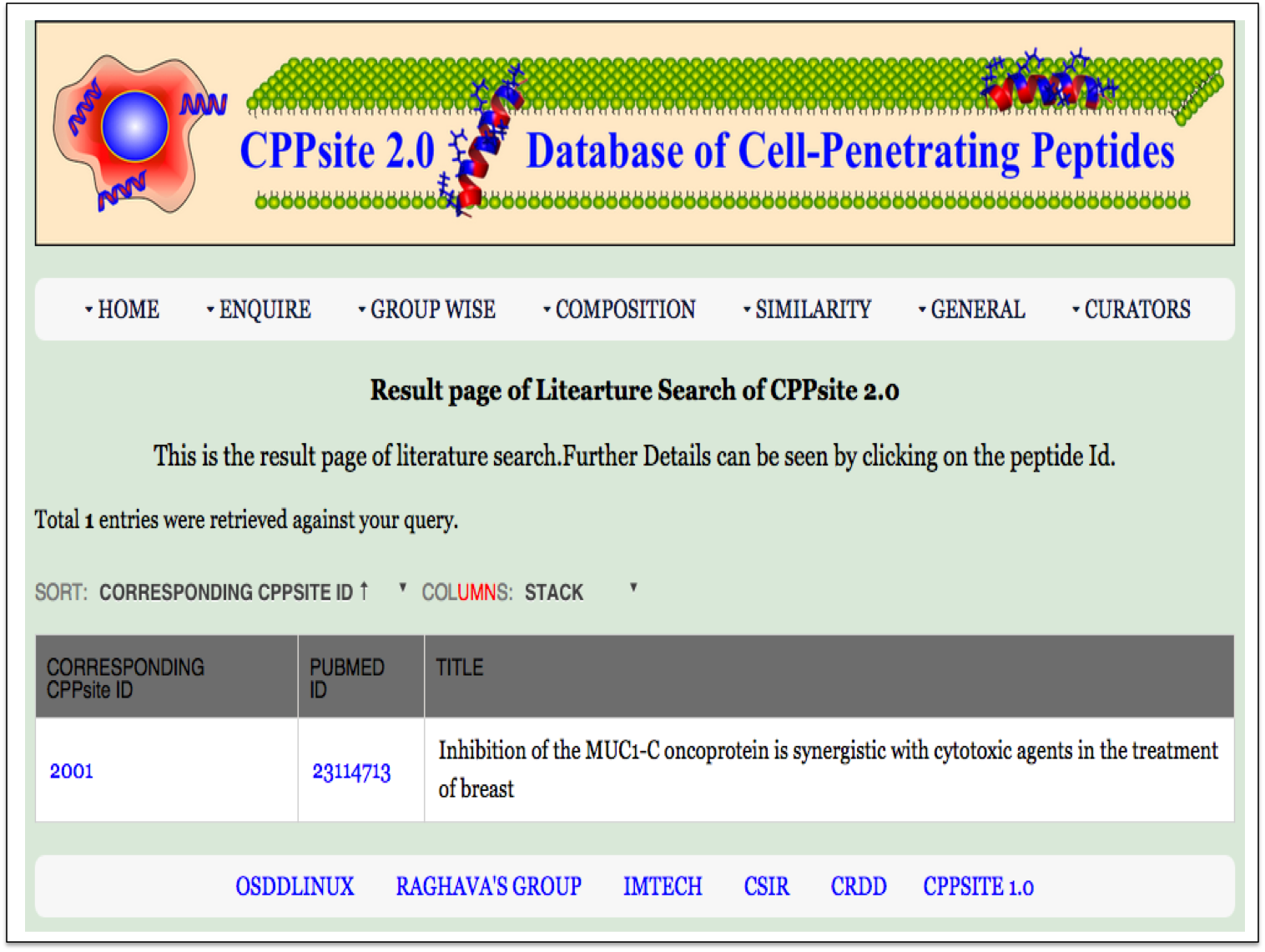
Literature Search This page provides the faciltiy to search CPPs by author name, title of the paper, abstract, keywords, etc. User can submit its query sequence and search it in the database. |
 |
| Result |
 |
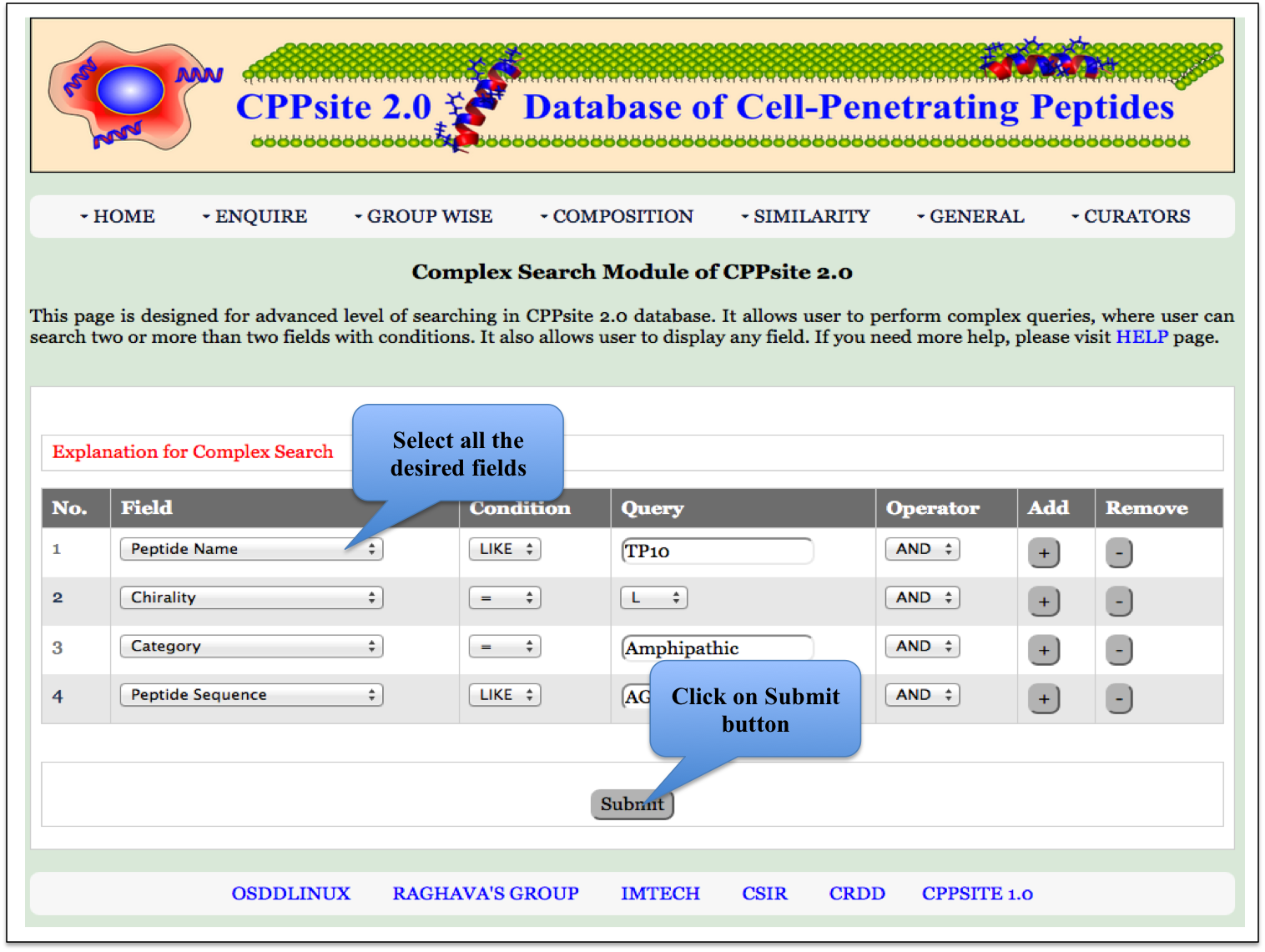
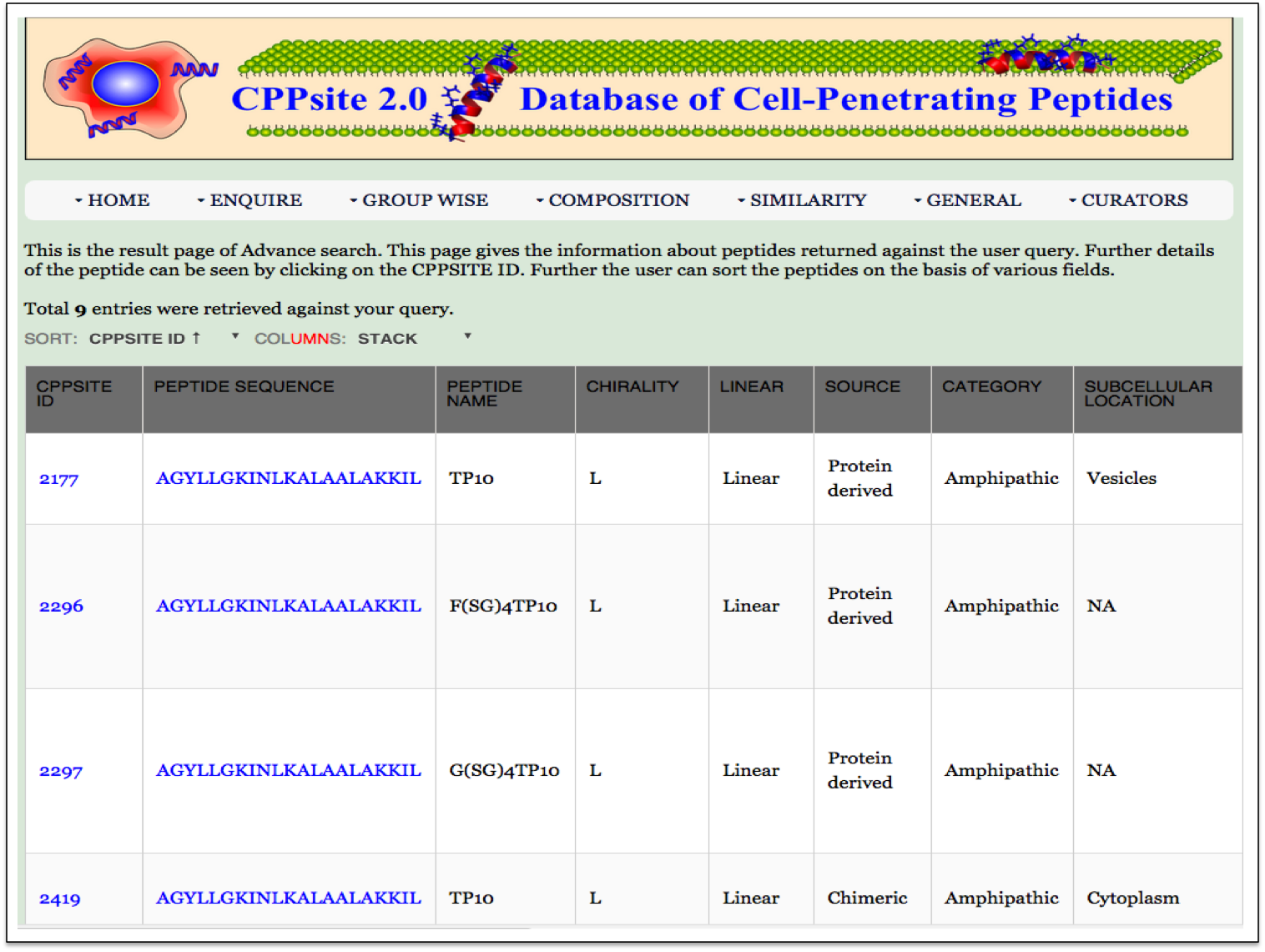
Complex Search This page provides the advanced search facility in CPPsite 2.0. User can browse the page by selecting two or more than two fileds. |
 |
| Result |
 |
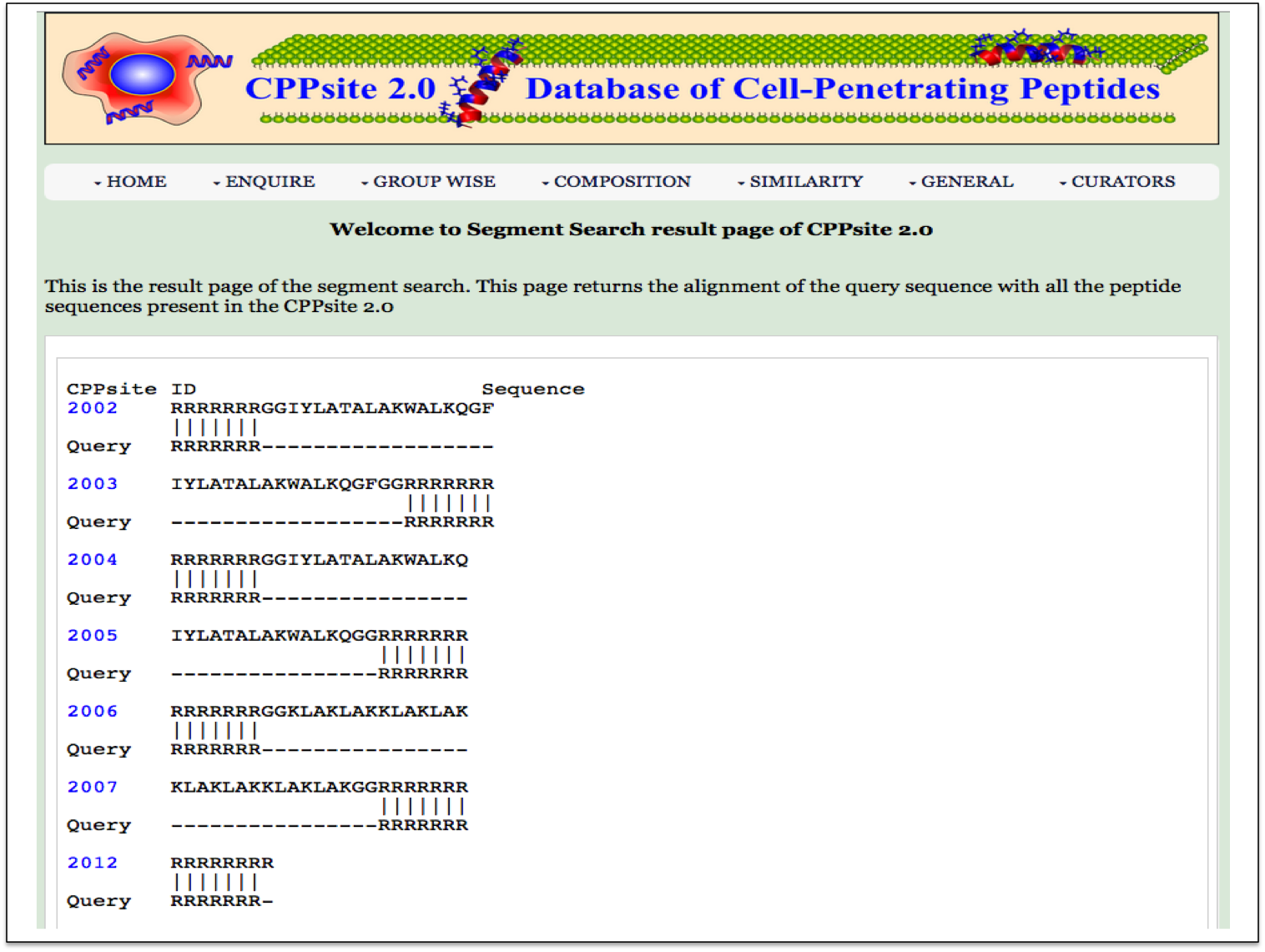
Segment Search This page provides facility to search his page returns the alignment of the query sequence with all the peptide sequences present in the CPPsite 2.0. |
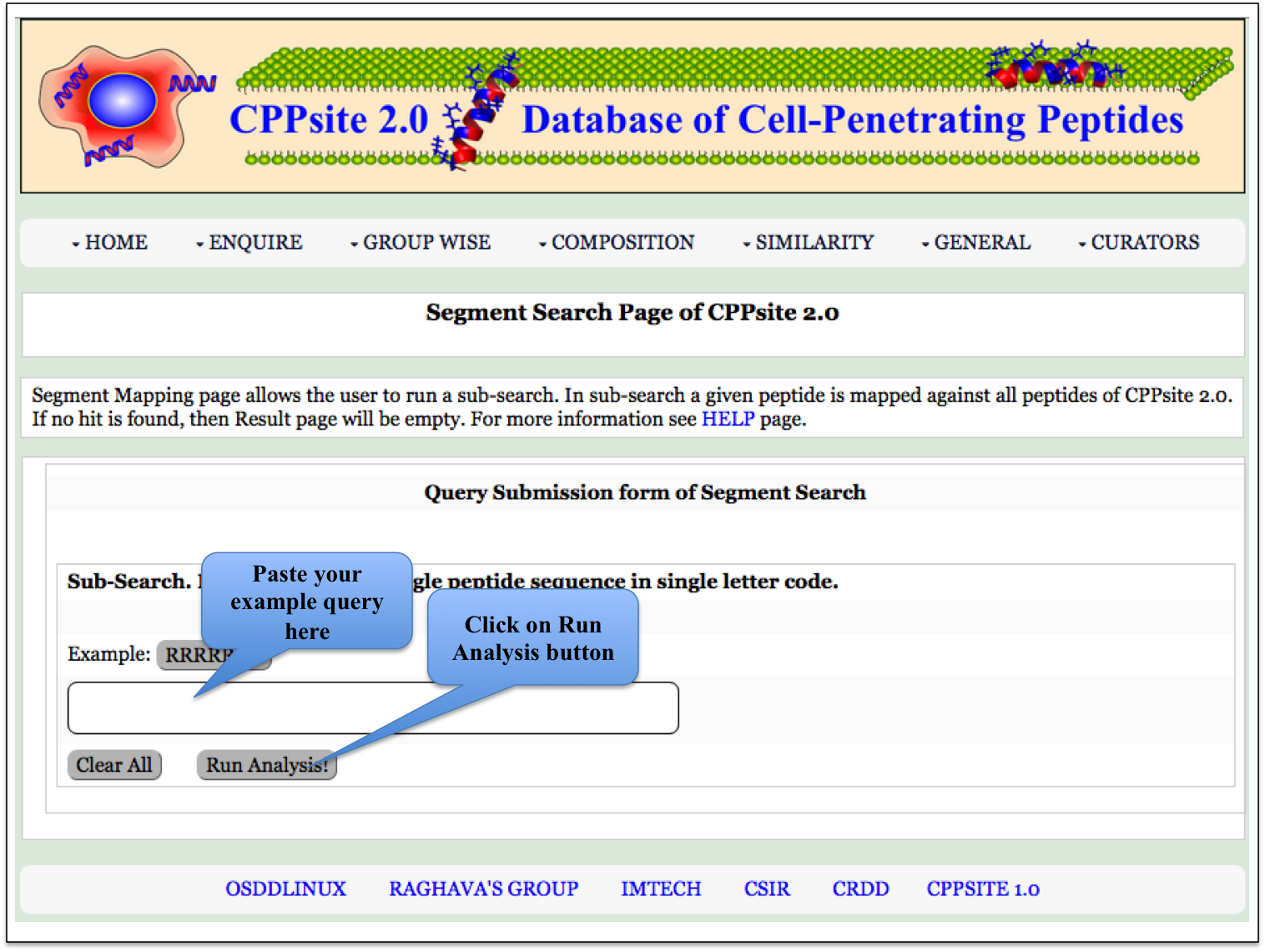 | Result |
 |
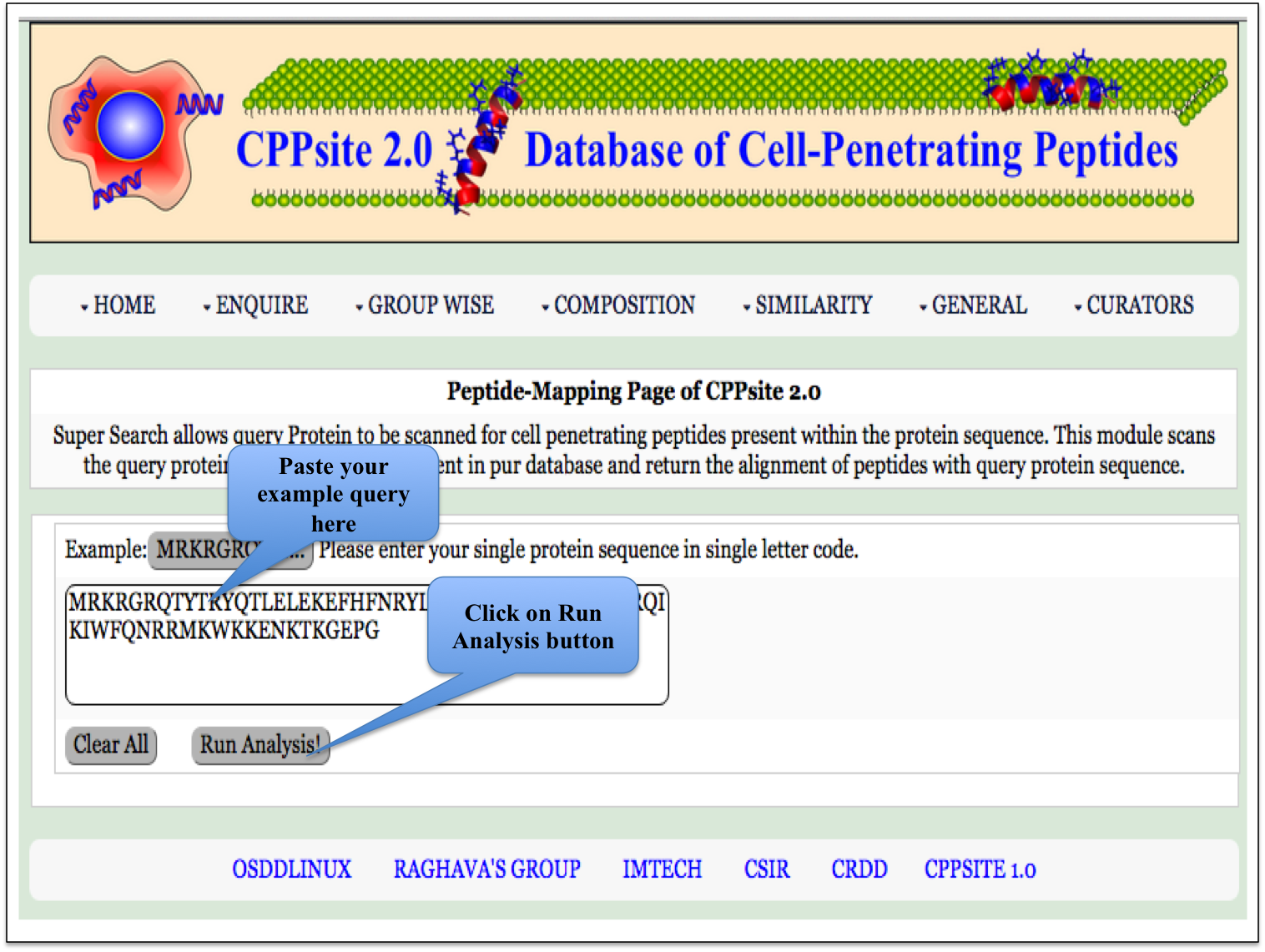
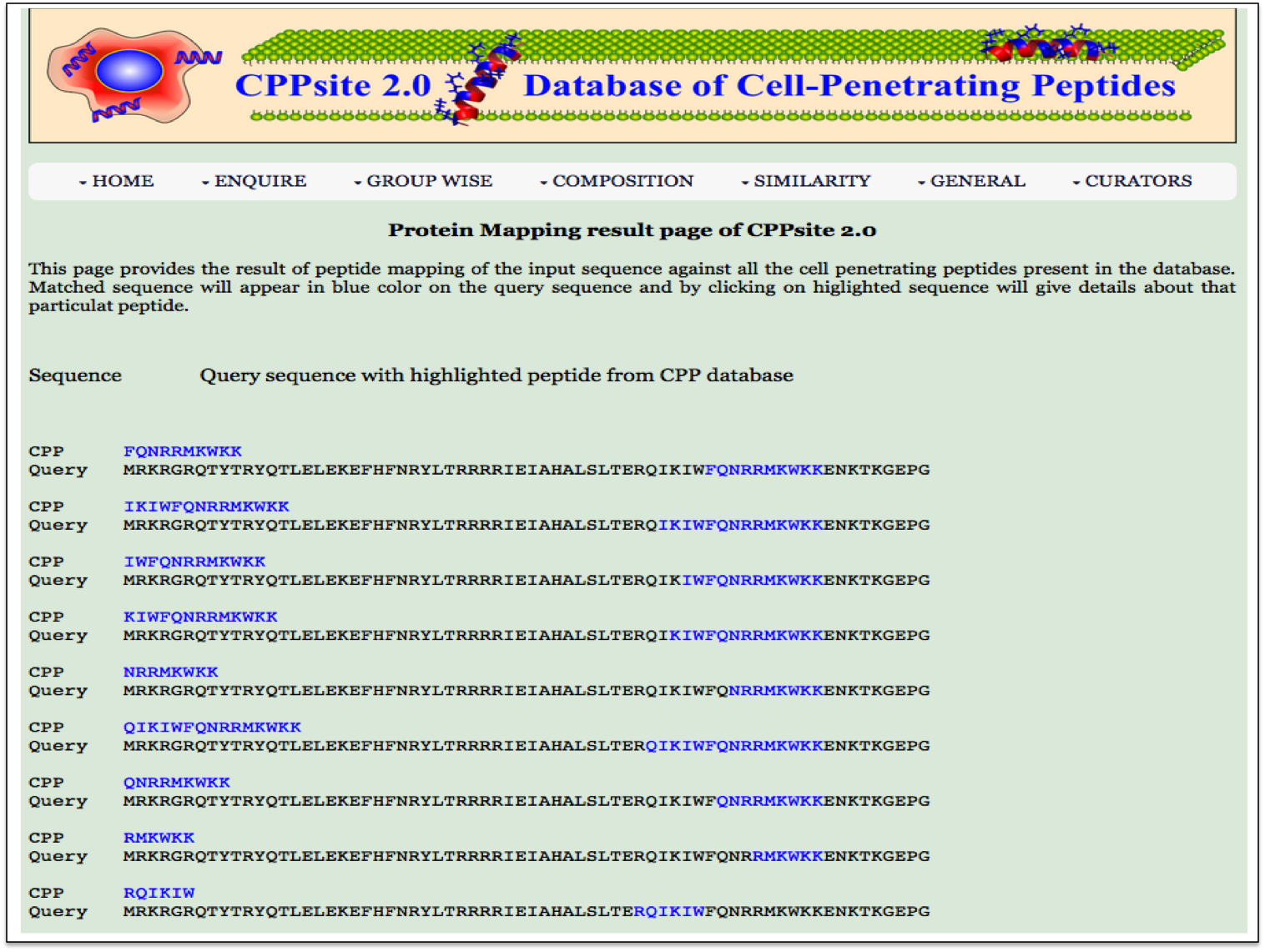
Peptide Mapping This page provides the facility of mapping the given query sequence with the cell penetrating peptide sequences present in the database CPPsite 2.0. |
 |
| Result |
 |
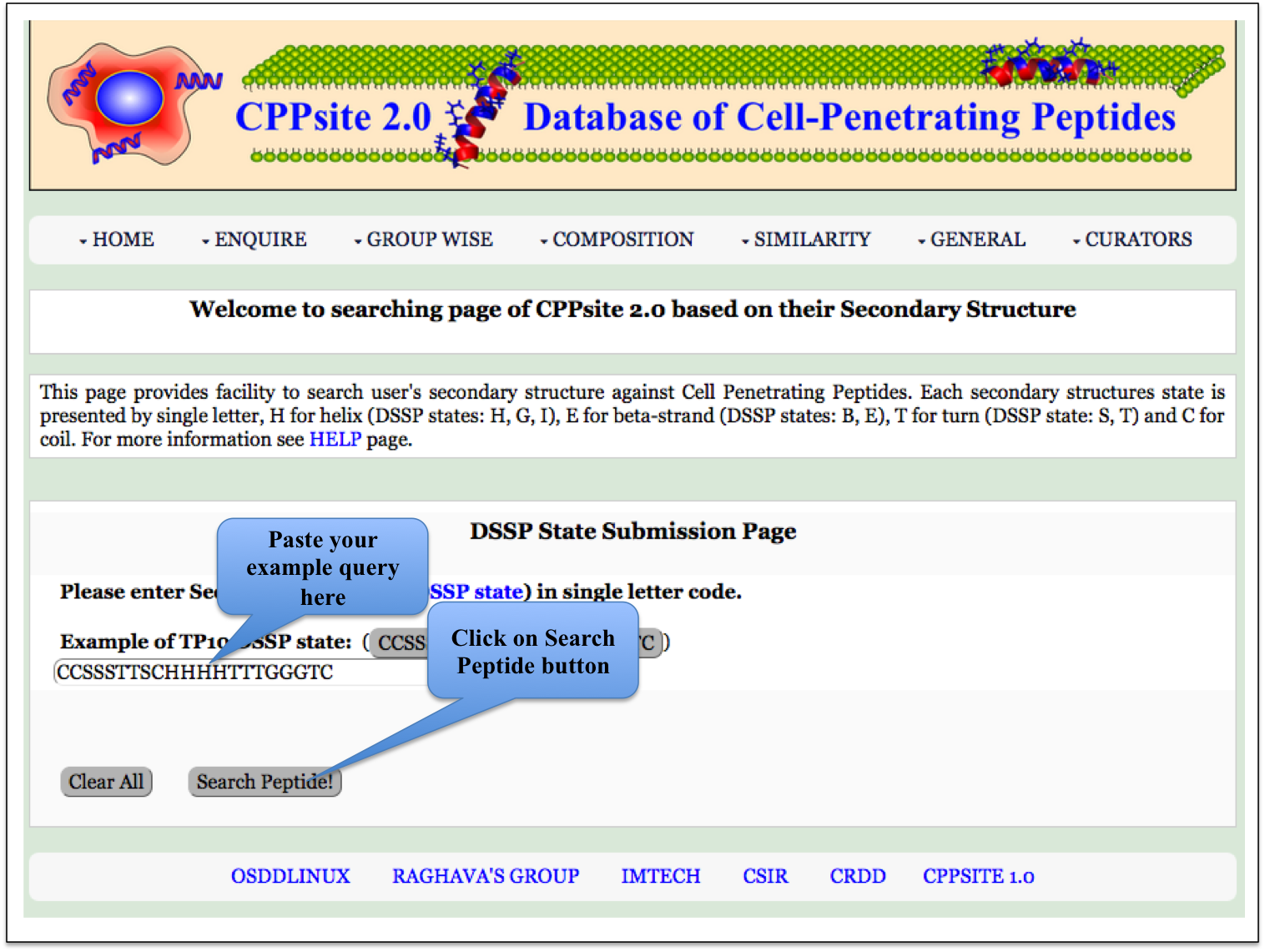
Secondary Structure This page provides the faciltiy to search user's secondary structure against CPPs. User can submit its query sequence and search it in the database. |
 |
| Result |
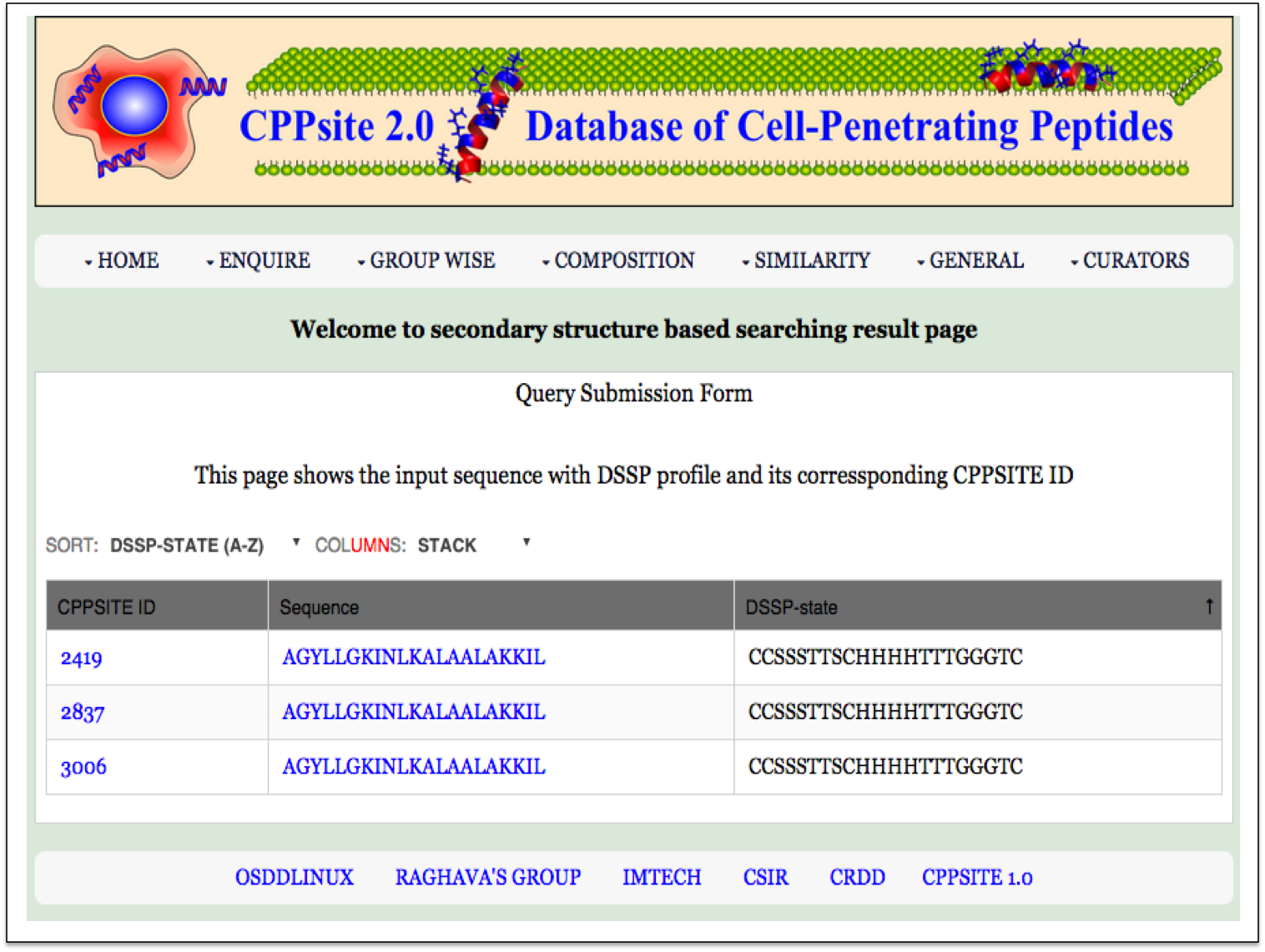 |
Group Wise |
Peptide Length This module displays the peptides of different length, present in the database. Clicking on number gives the details about the type of peptide selected. |
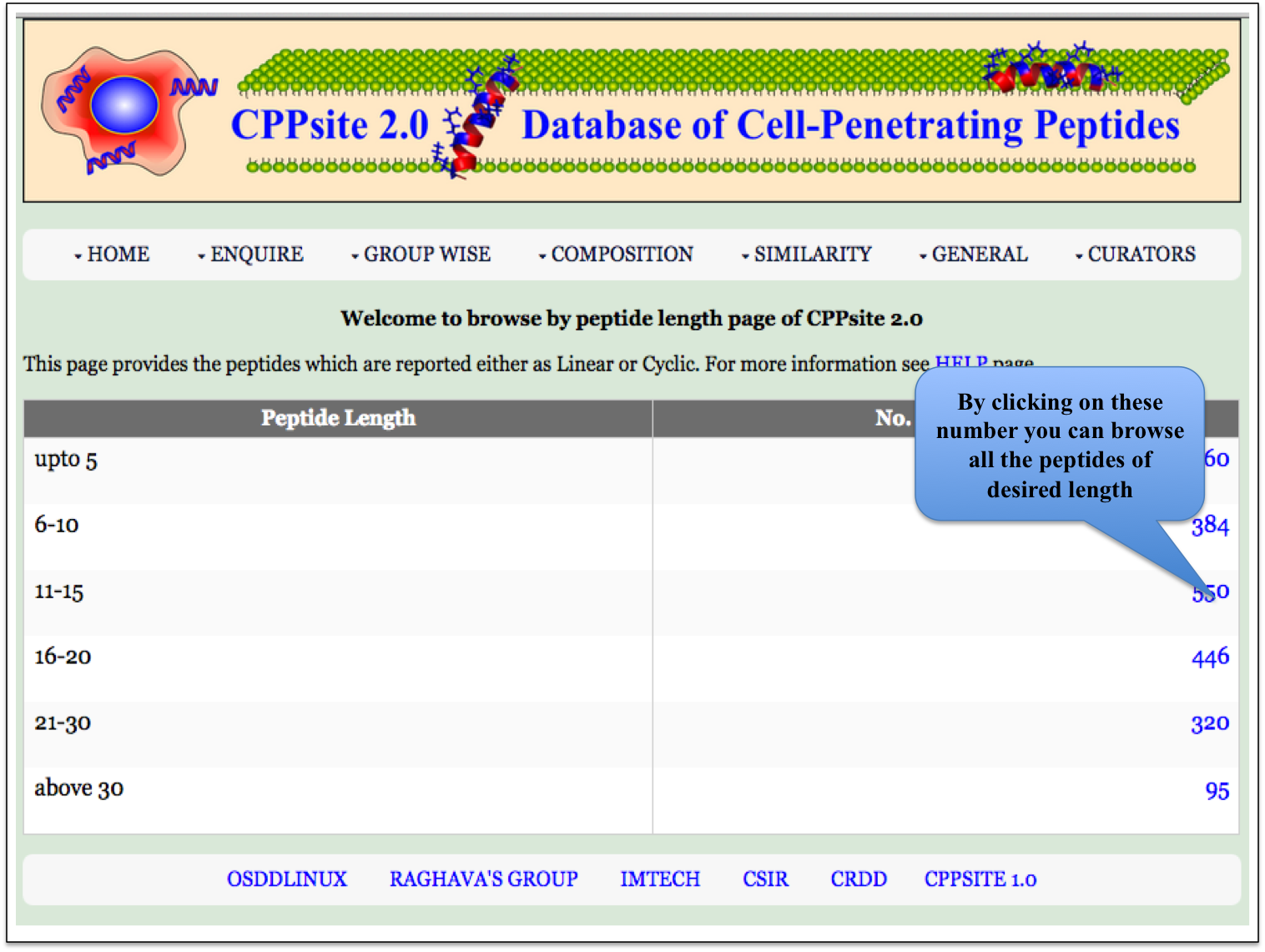 |
| Result |
 |
Category This module displays the category of peptides present in the database. Clicking on number gives the details about the type of peptide selected. |
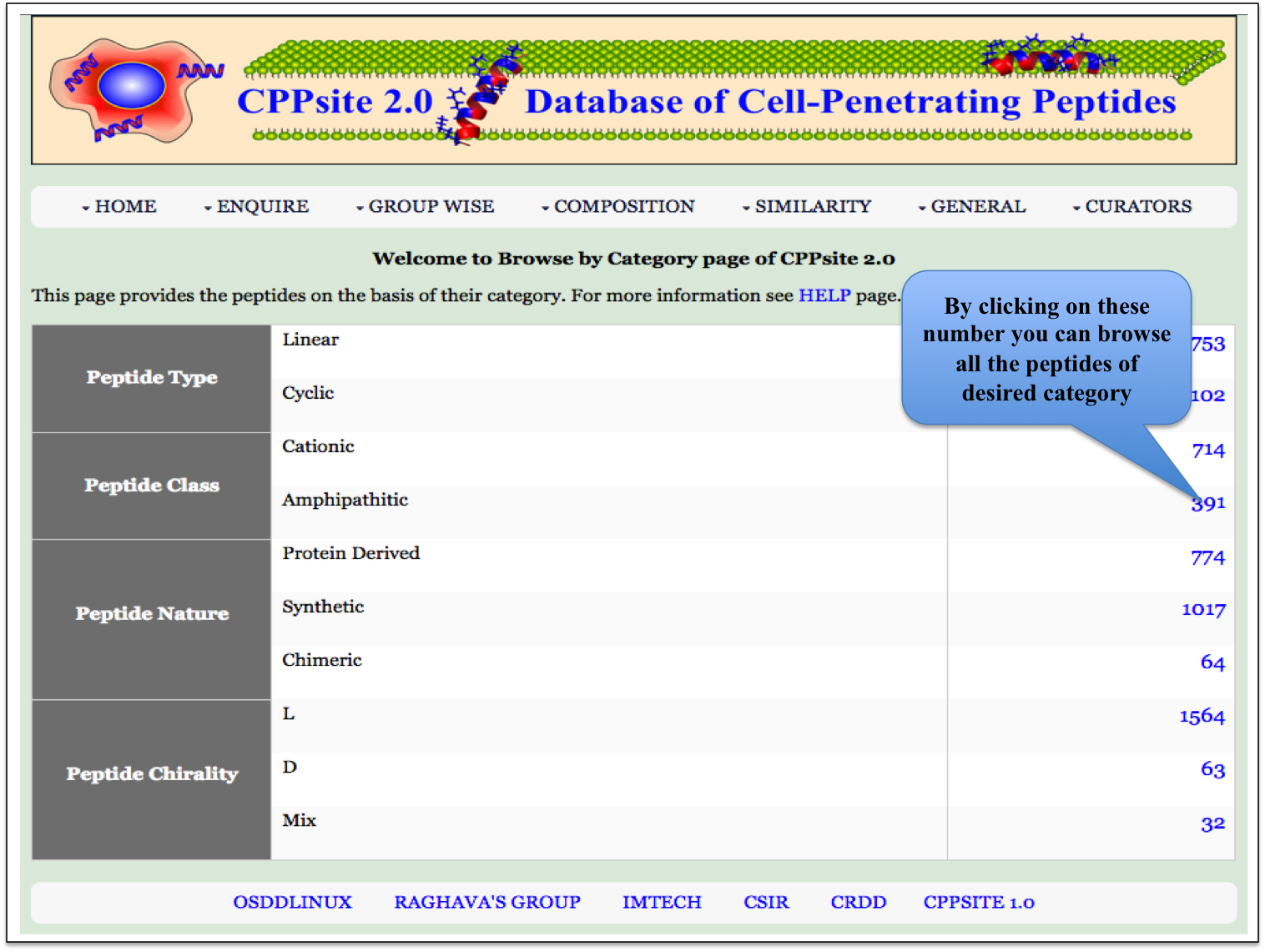 |
| Result |
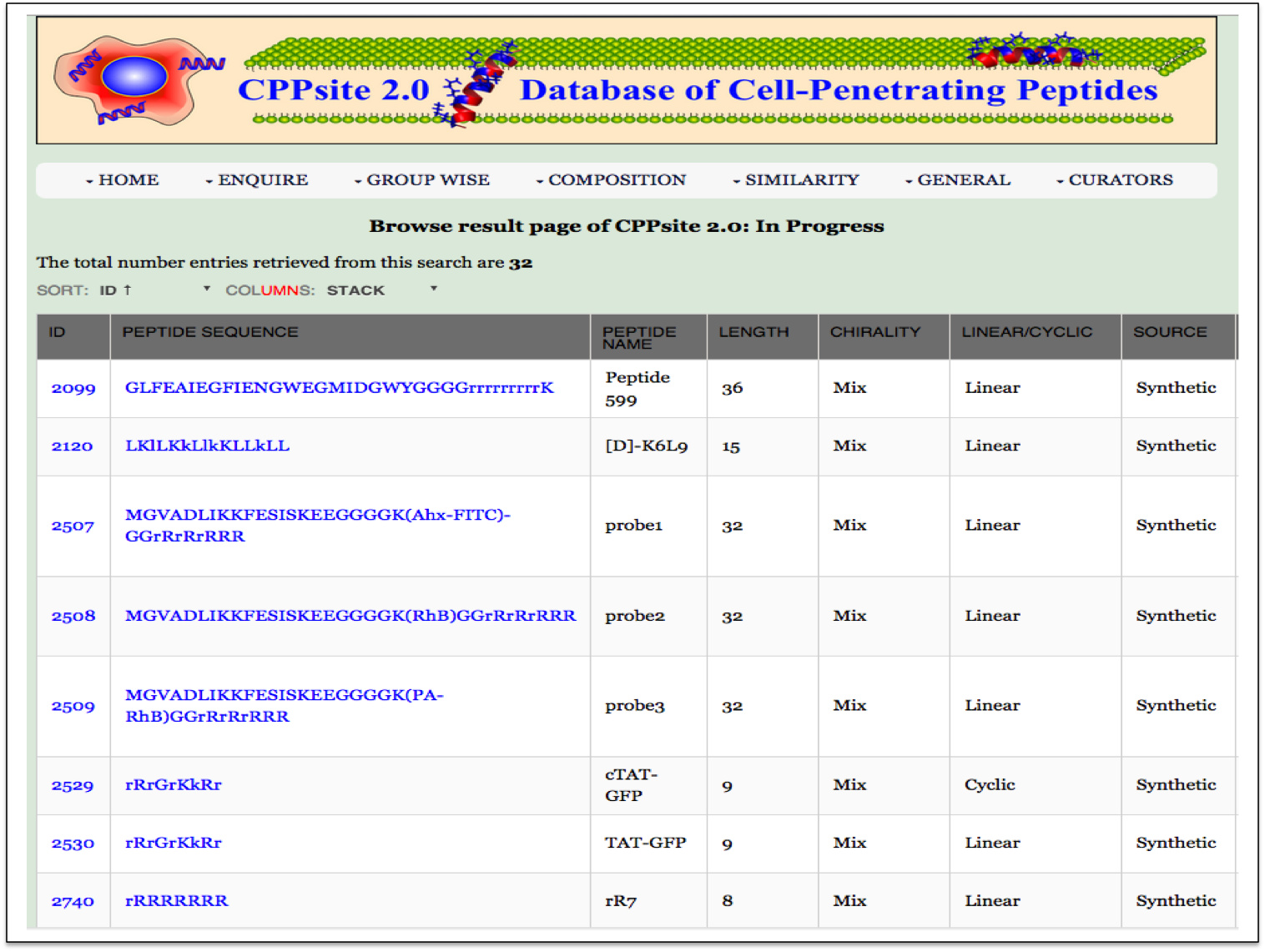 |
Chemical Modification This page provides the distribution of different modifications (Terminal modifications, Various chemical modifications) present in the peptides of CPPsite 2.0 database. |
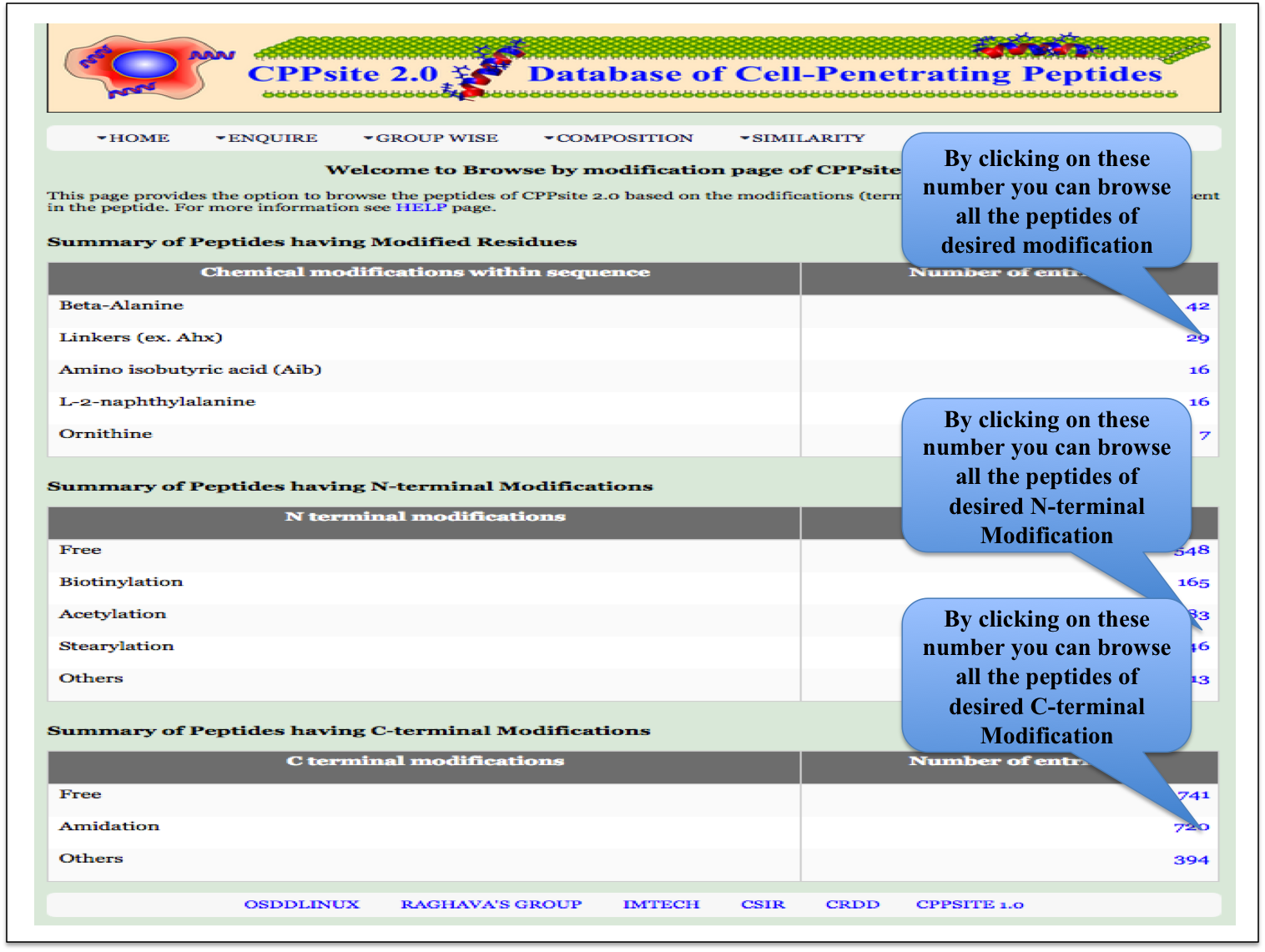 |
| Result |
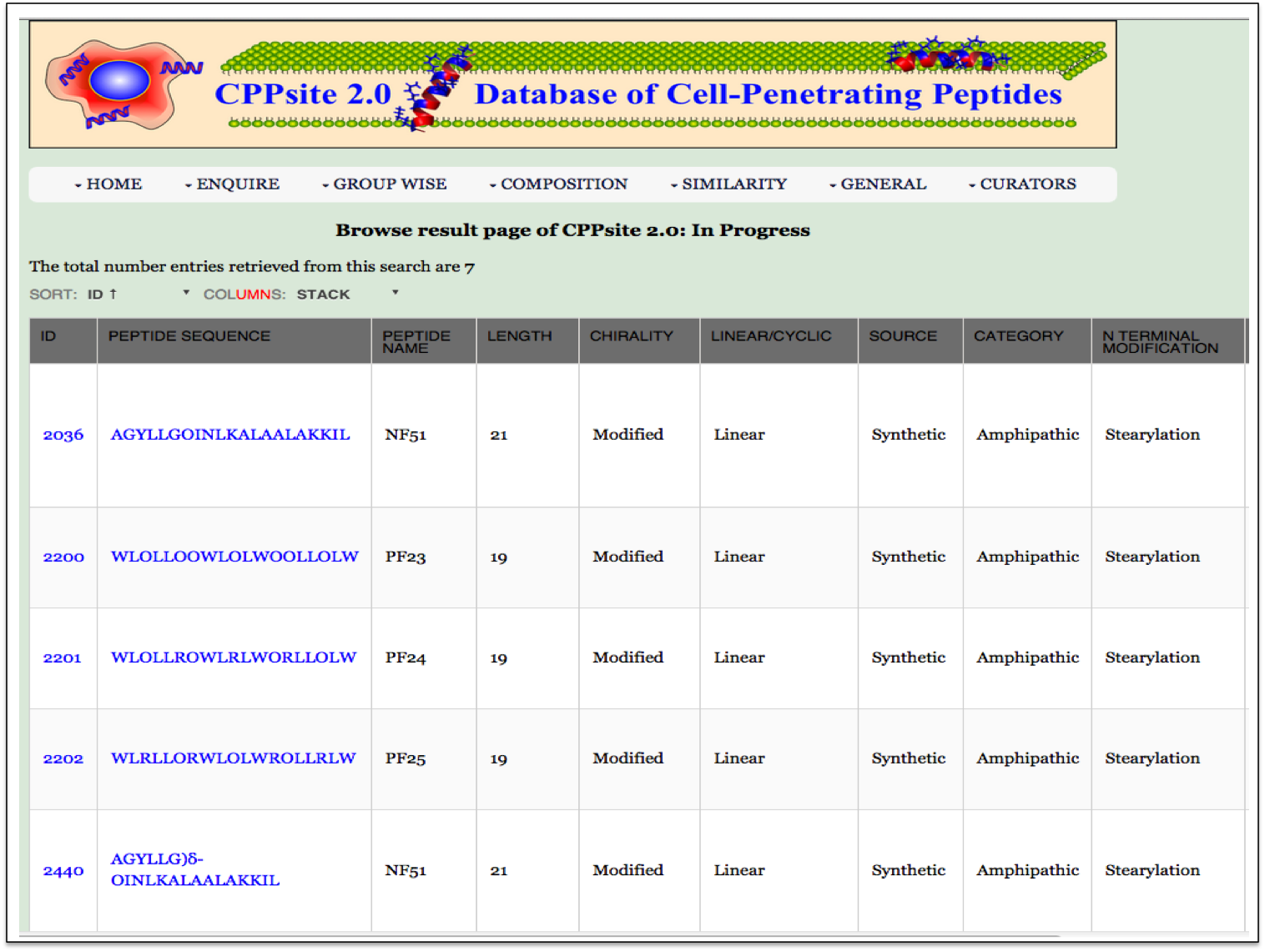 |
Cargo This module displays the types of cargoes delivered with the CPPs. Clicking on number gives all the details about the particular cargo present in the database CPPsite 2.0. |
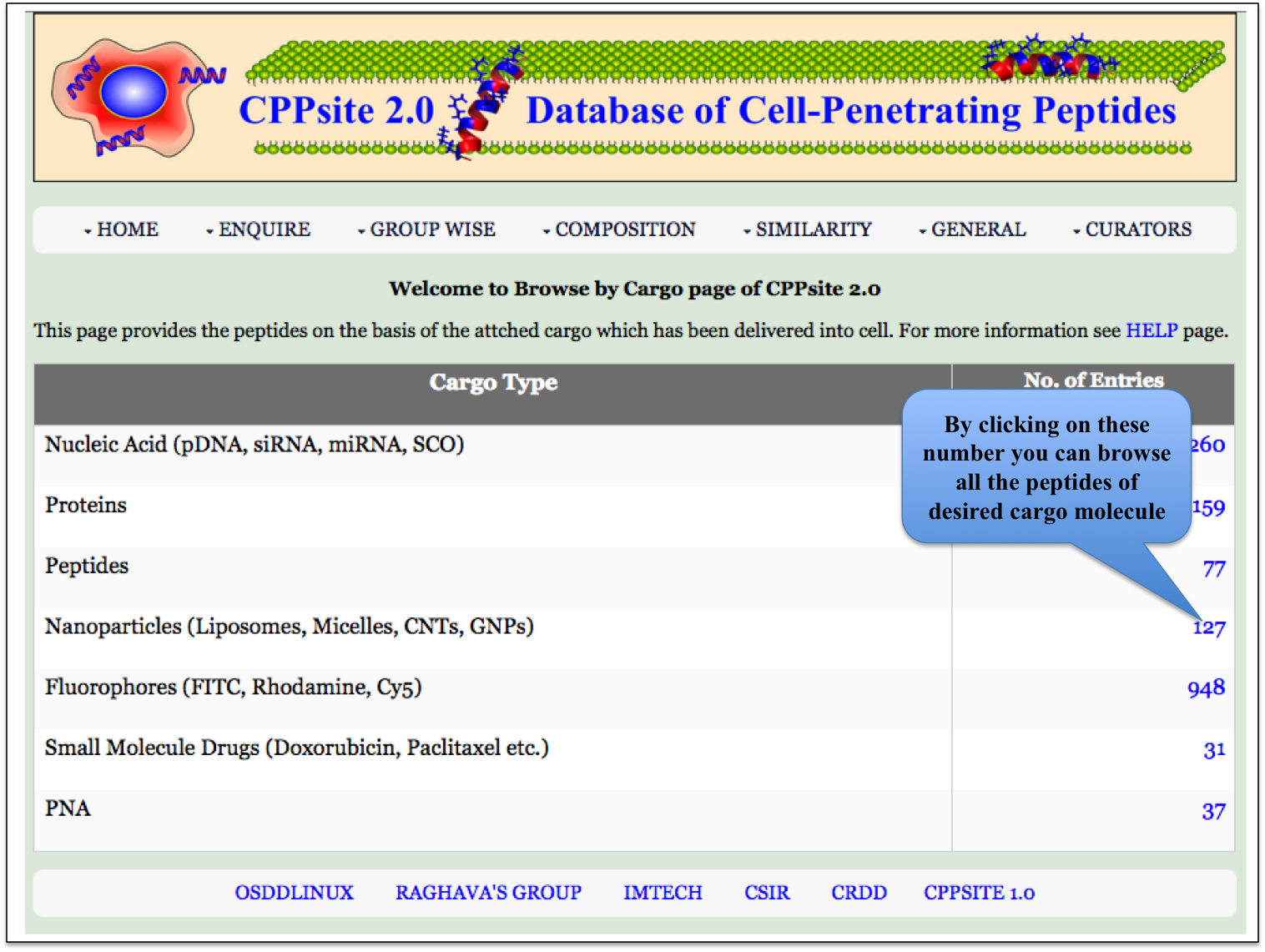 |
| Result |
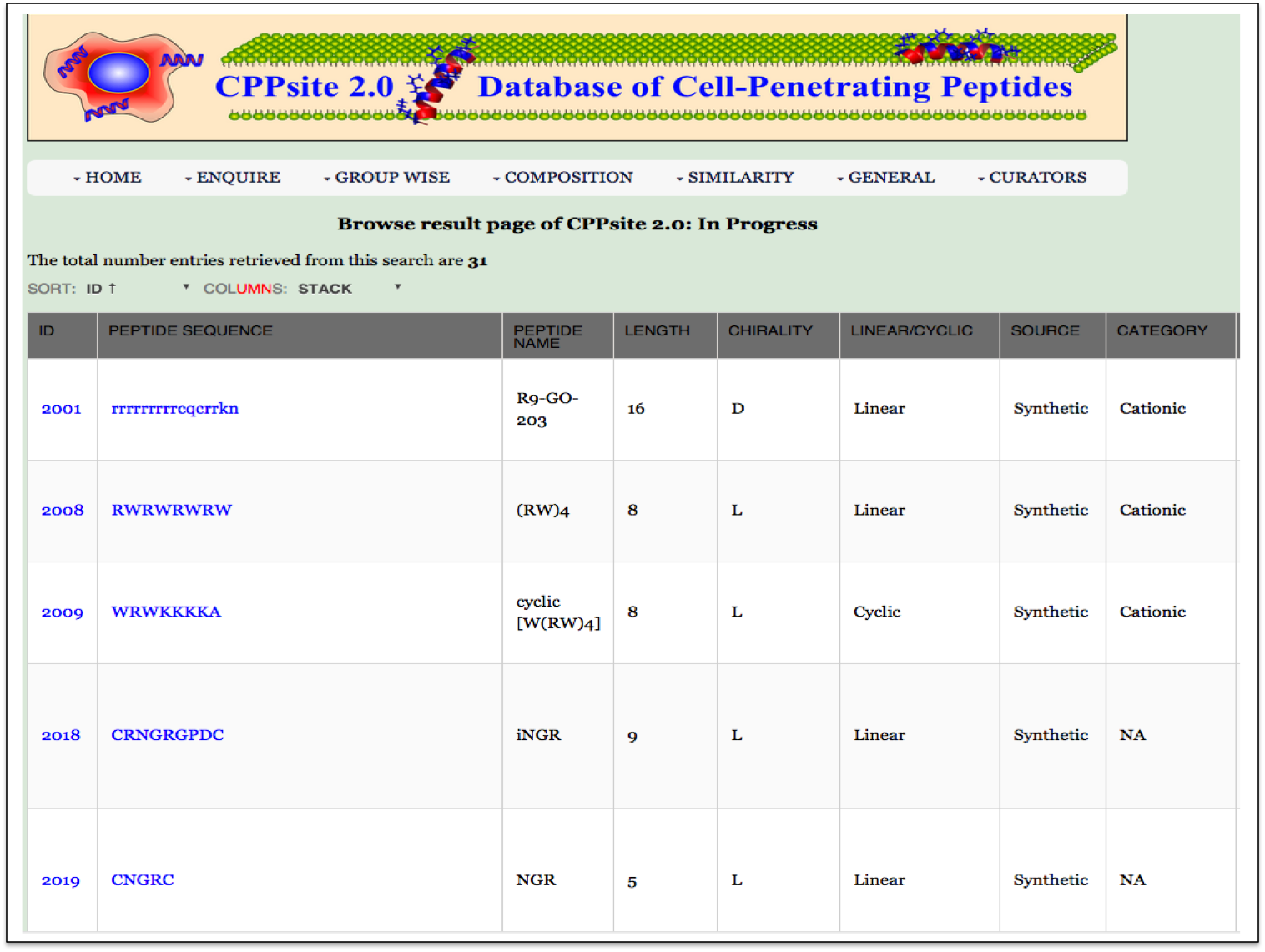 |
Composition |
AA Frequency Here the user can browse the peptides based on the frequency of amino acid. |
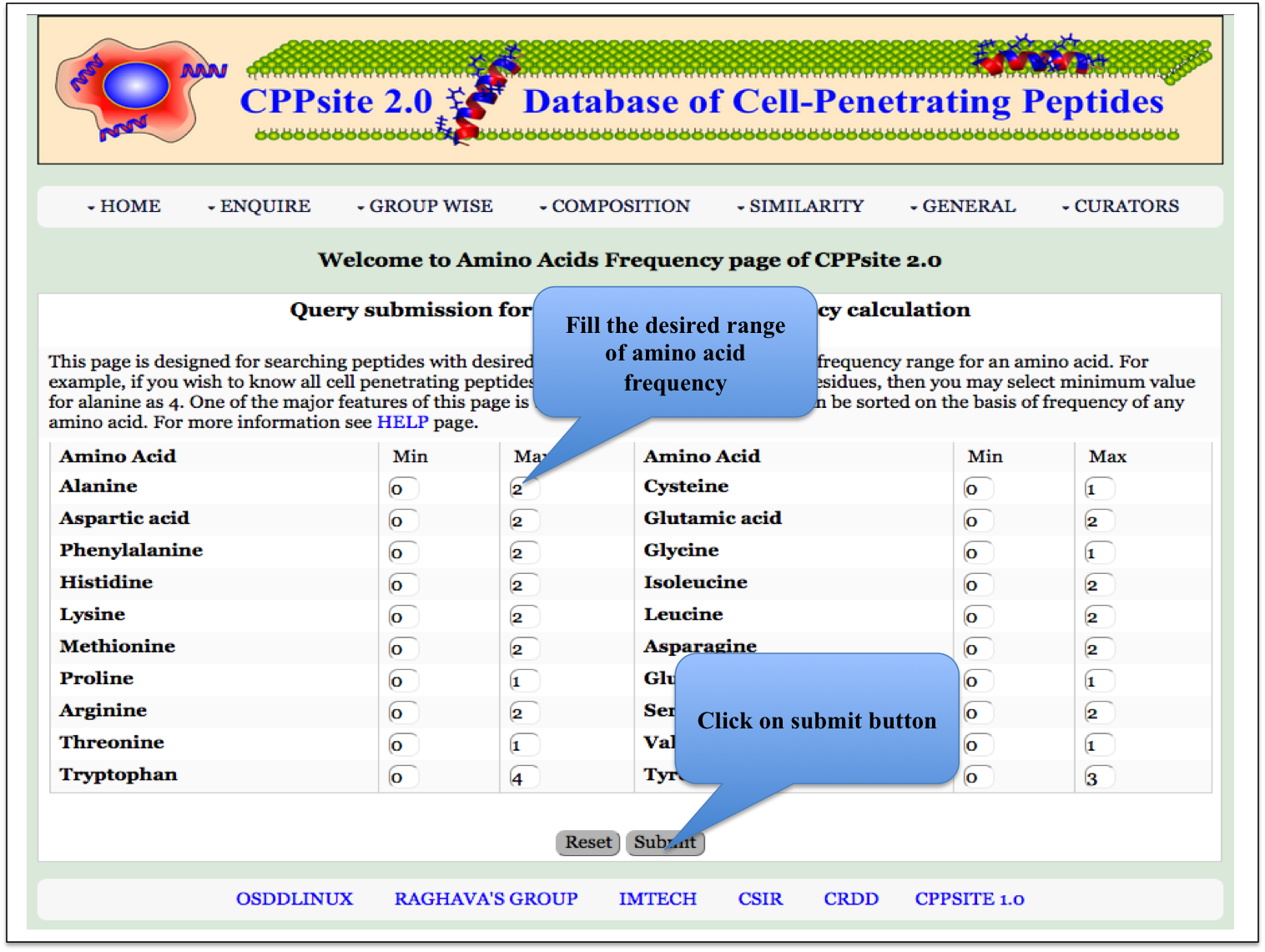 |
| Result |
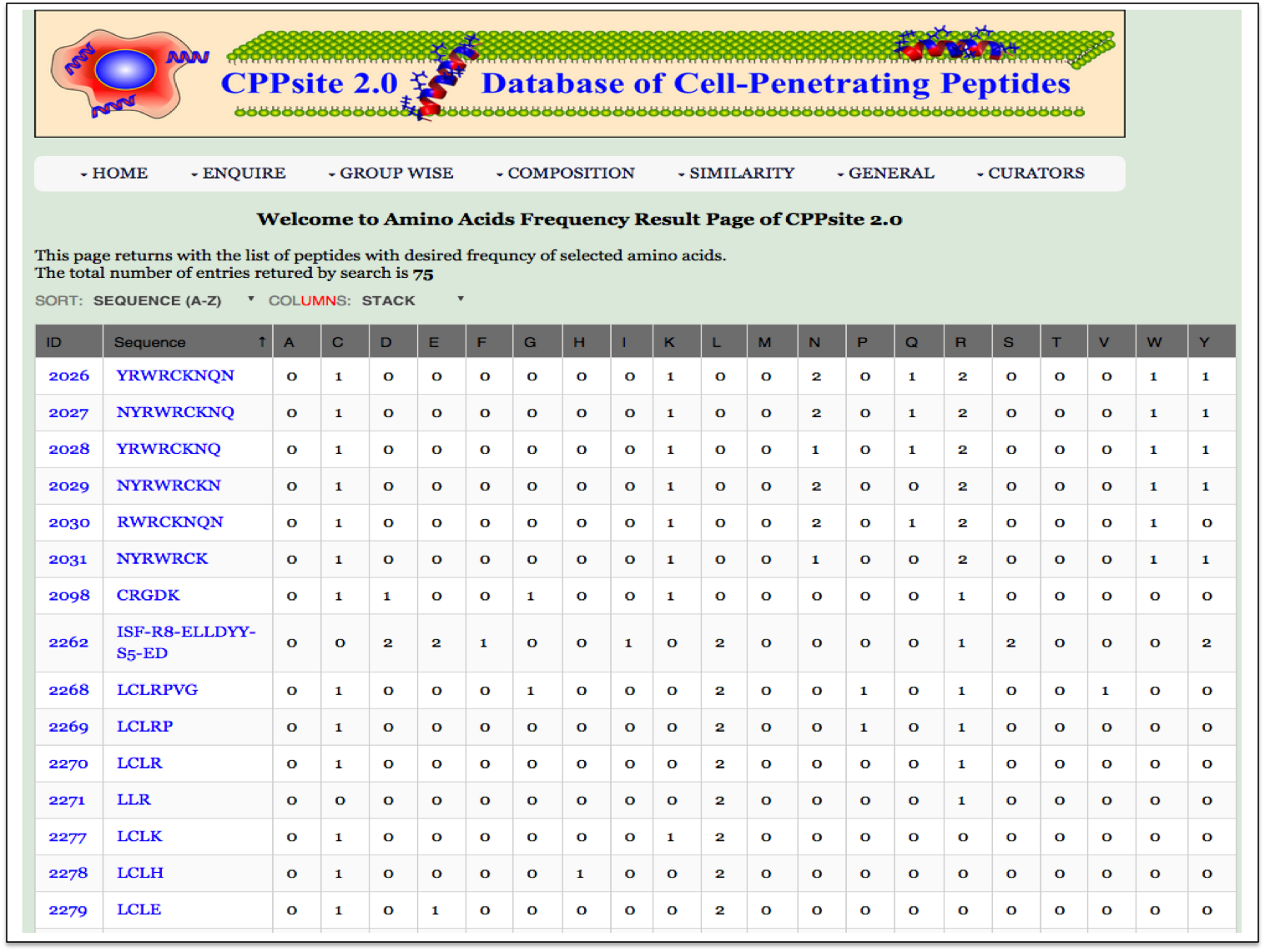 |
PP Frequency Here the user can browse the peptides based on the frequency of physiochemical properties. |
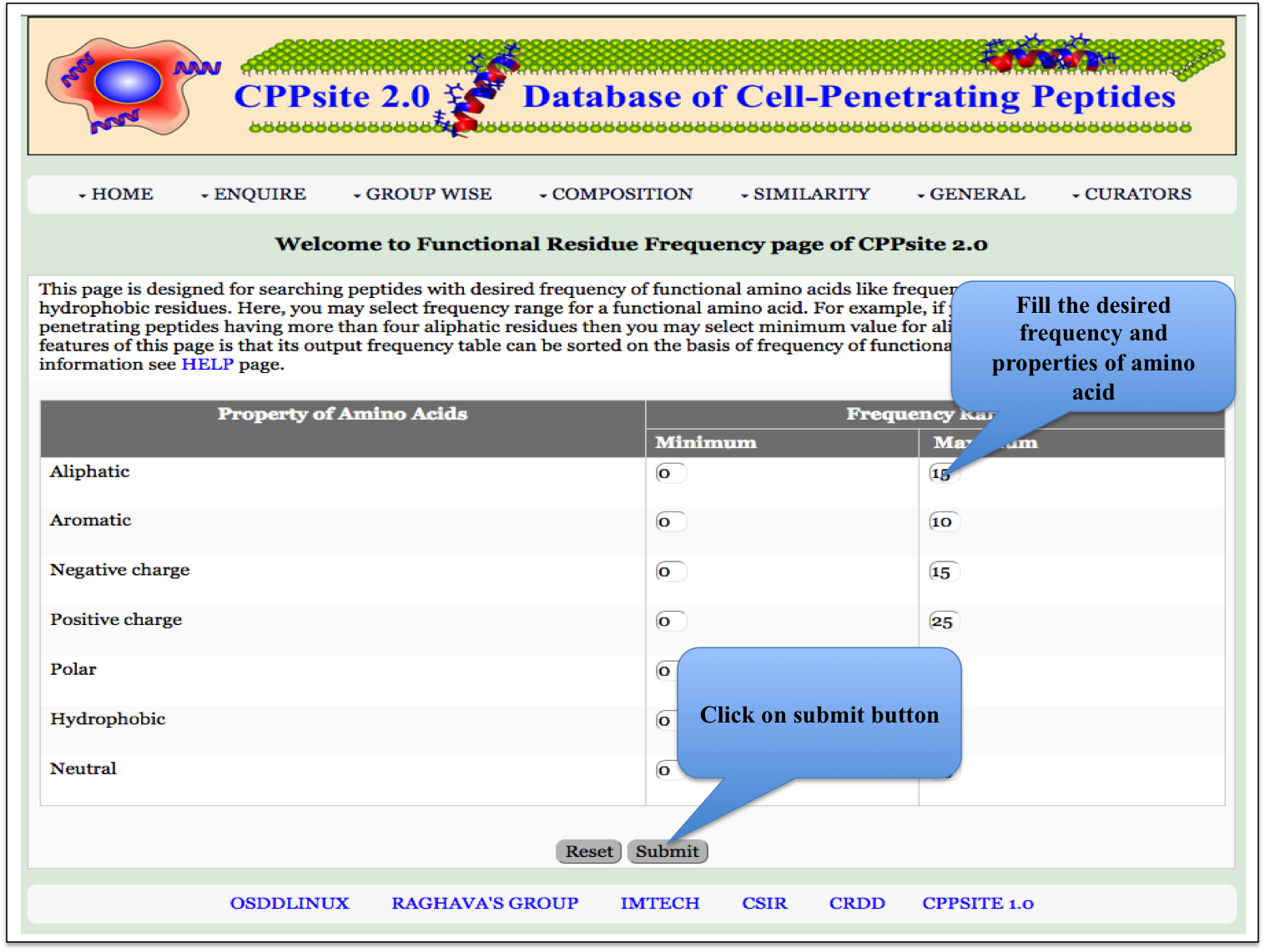 |
| Result |
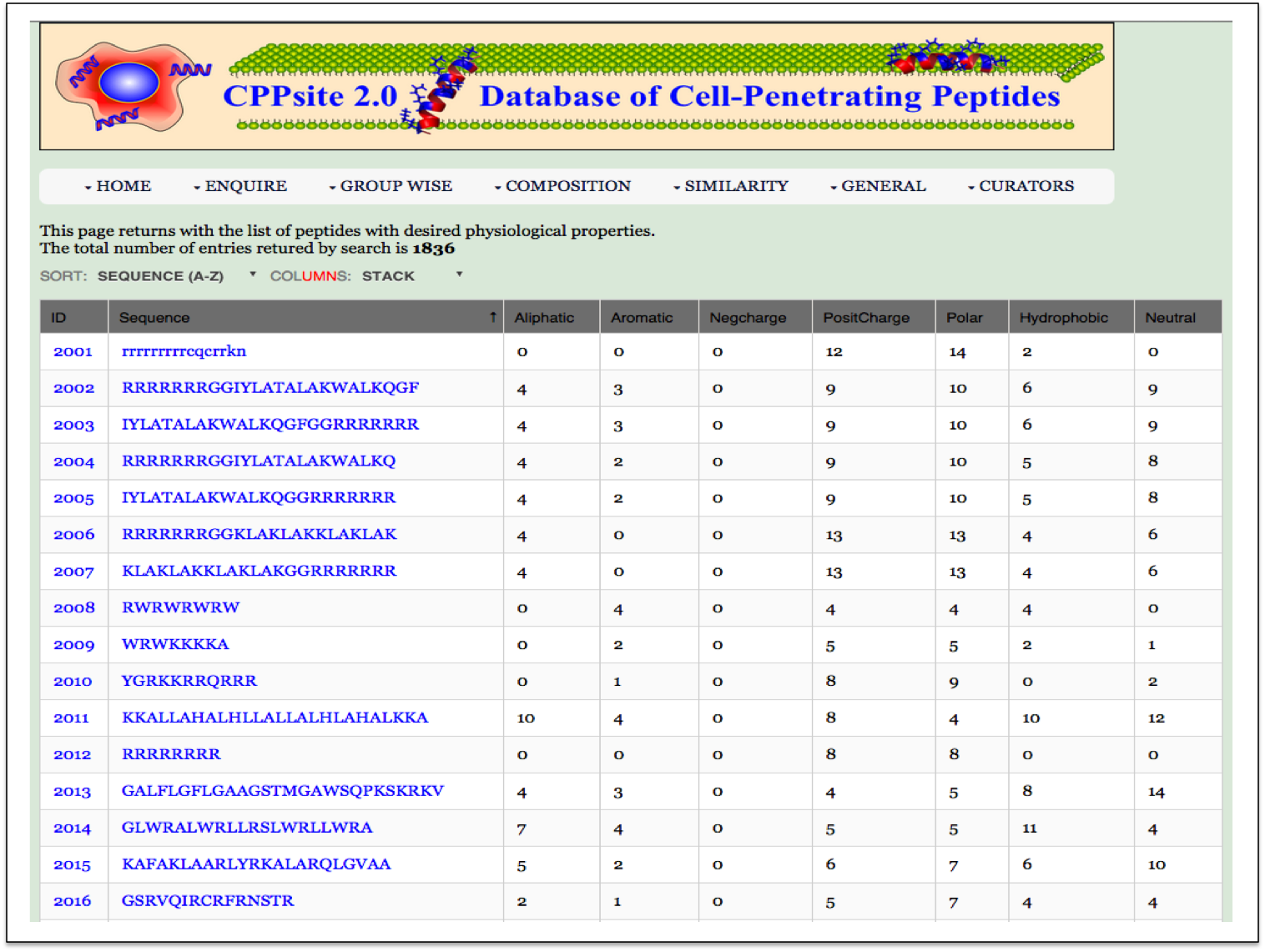 |
AA Composition Here the user can browse the peptides with desired amino acid composition. |
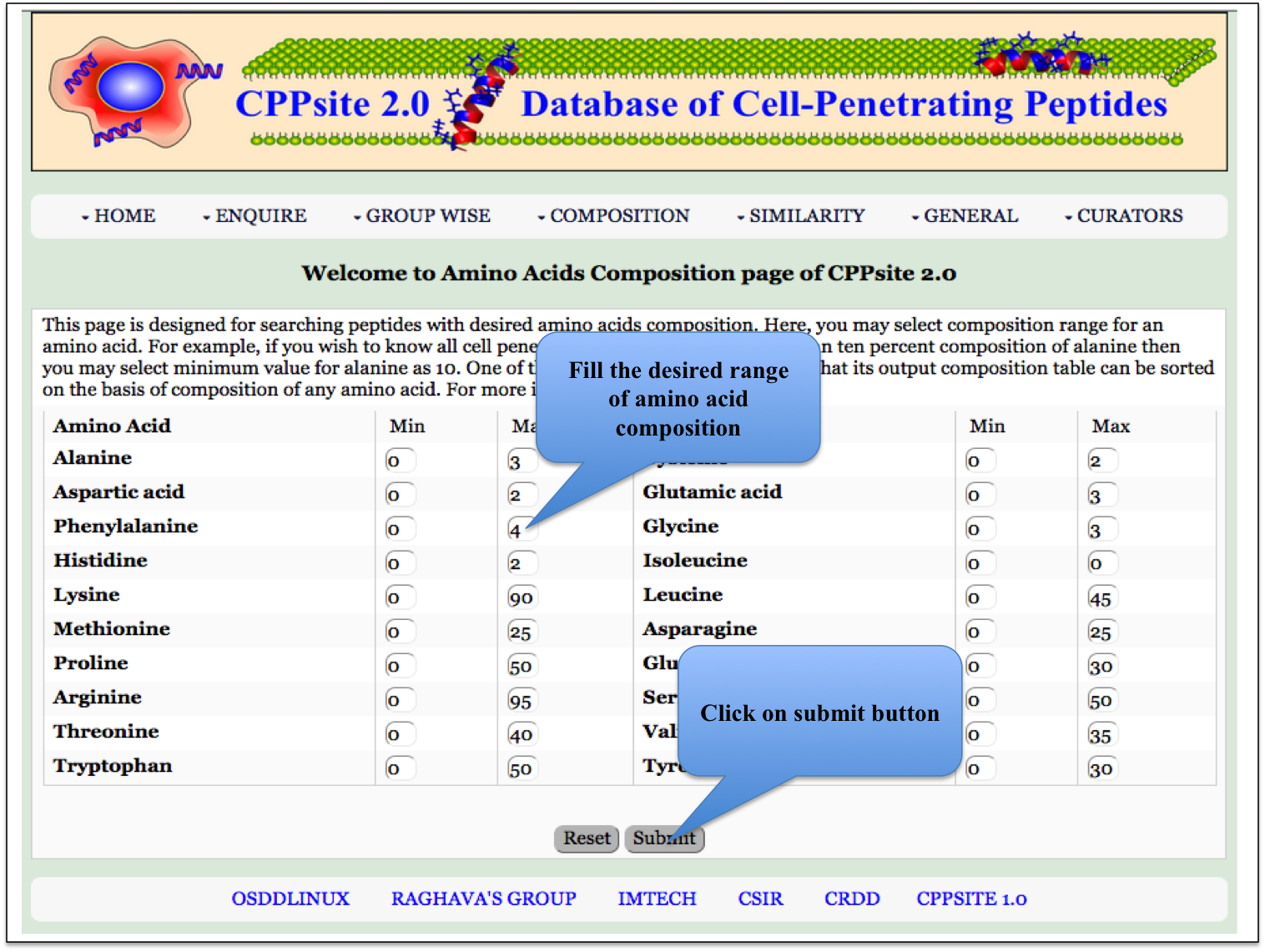 |
| Result |
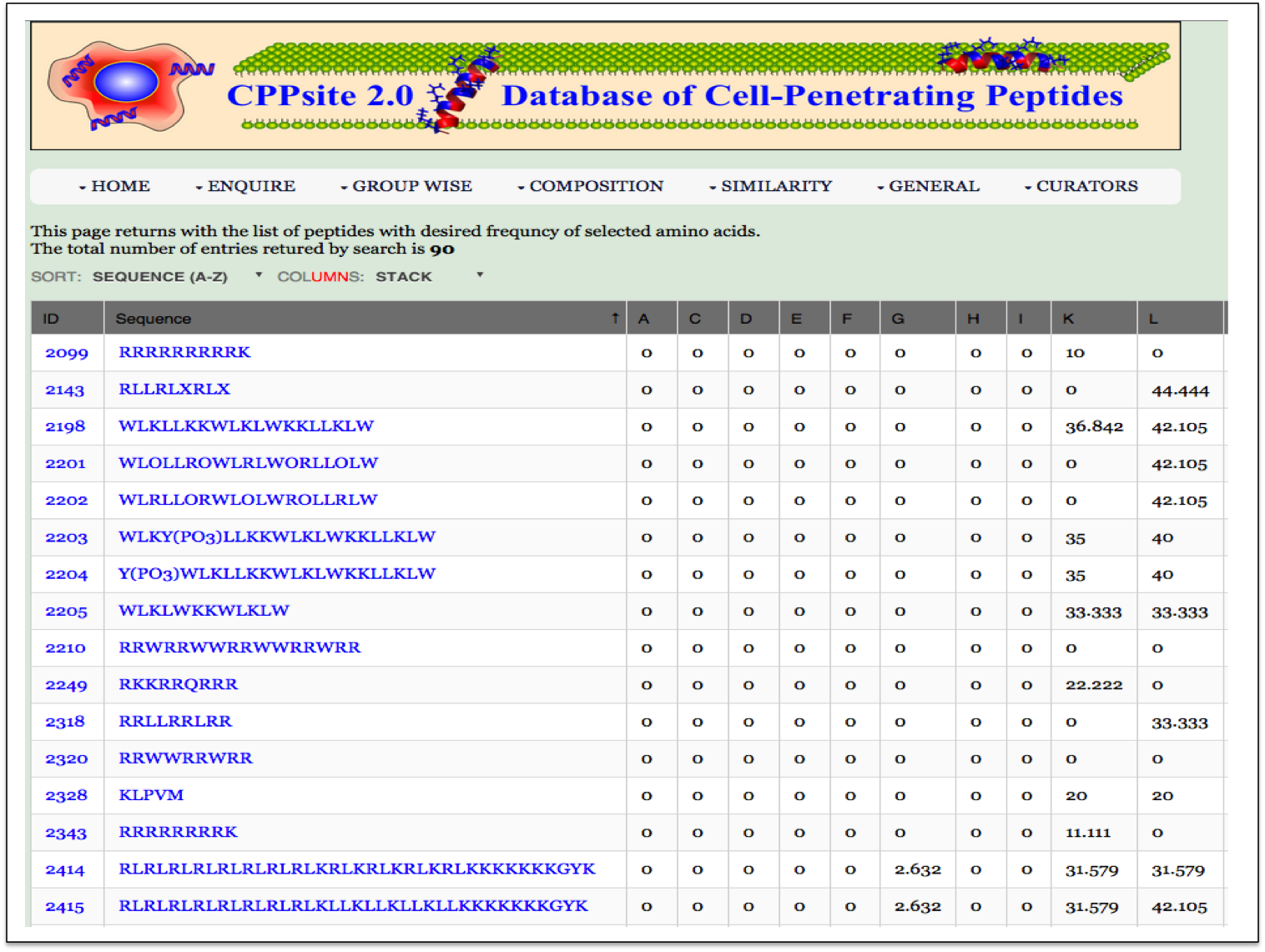 |
PP Composition Here the user can browse the peptides with desired physiochemical composition. |
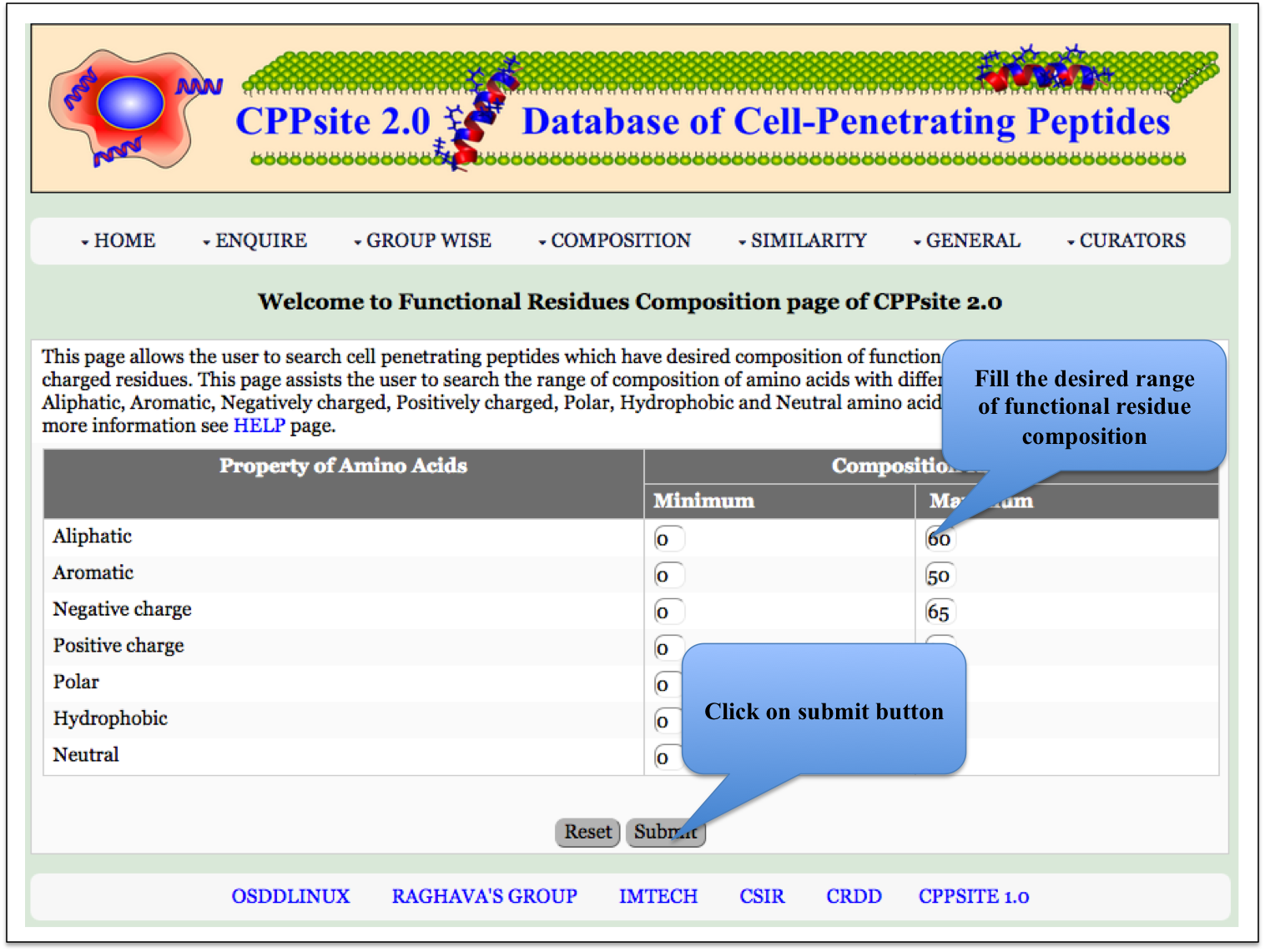 |
| Result |
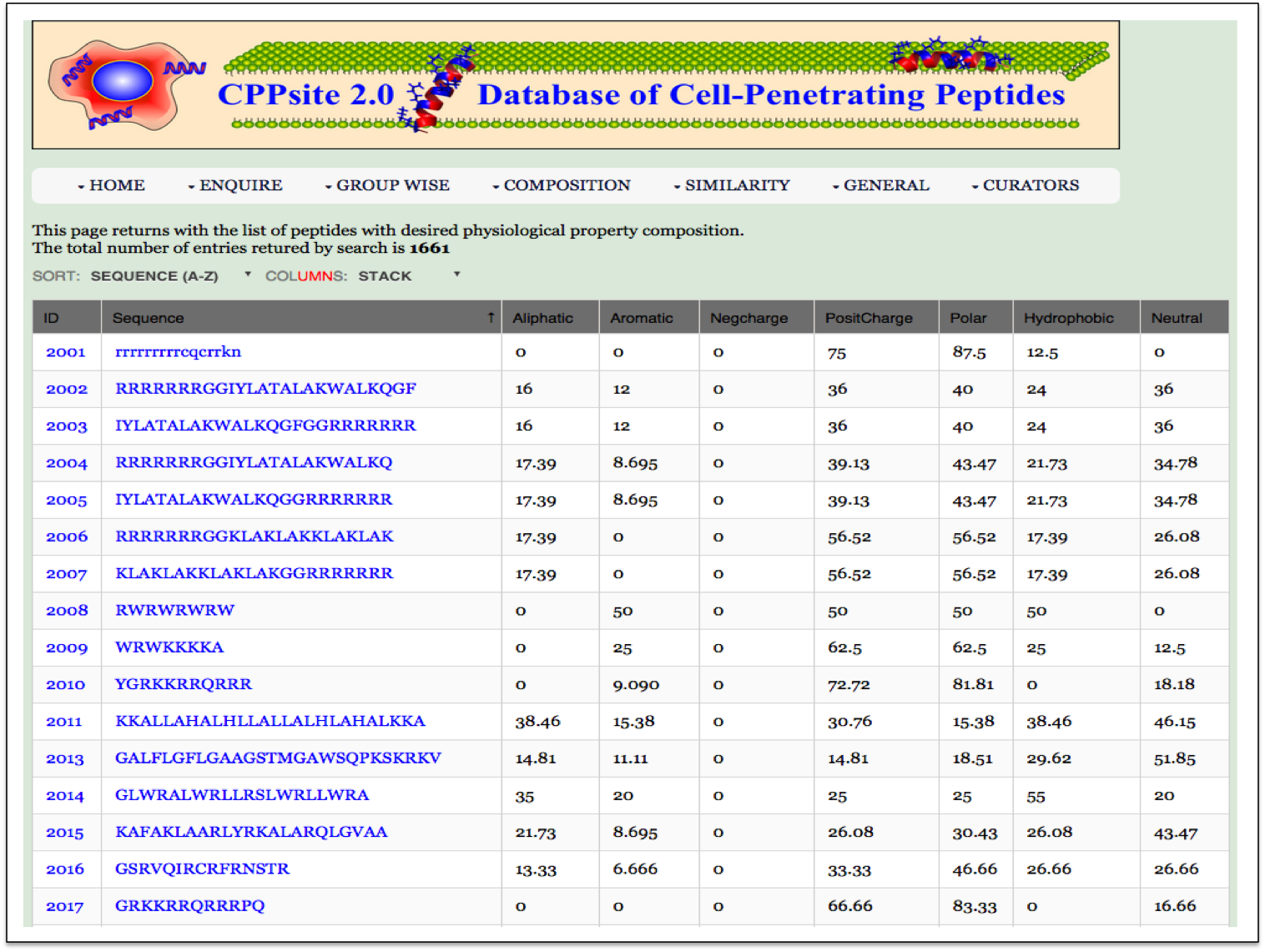 |
SS Composition This page assits the user to search cell penetrating peptides based on their secondary structure composition. |
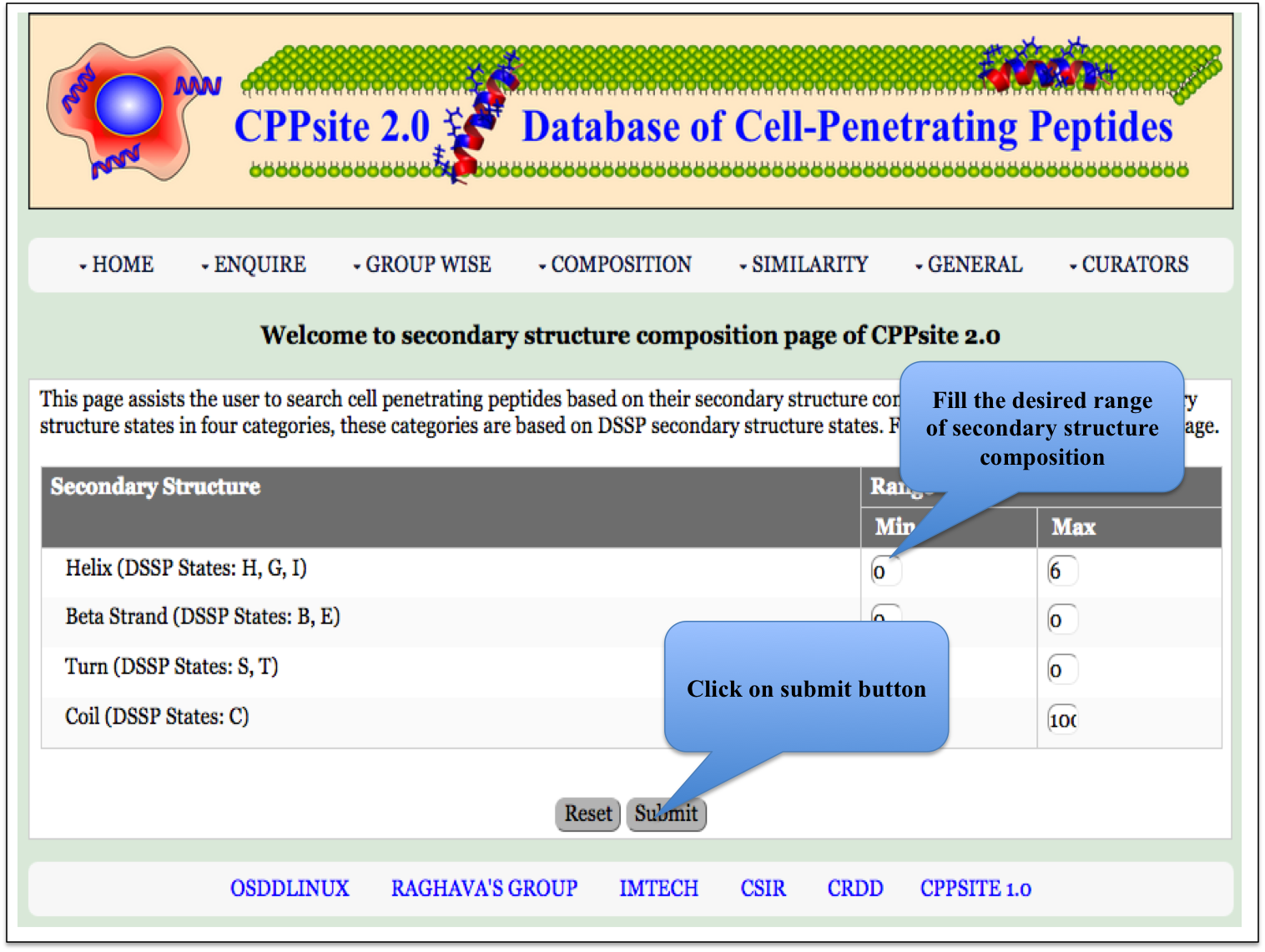 |
| Result |
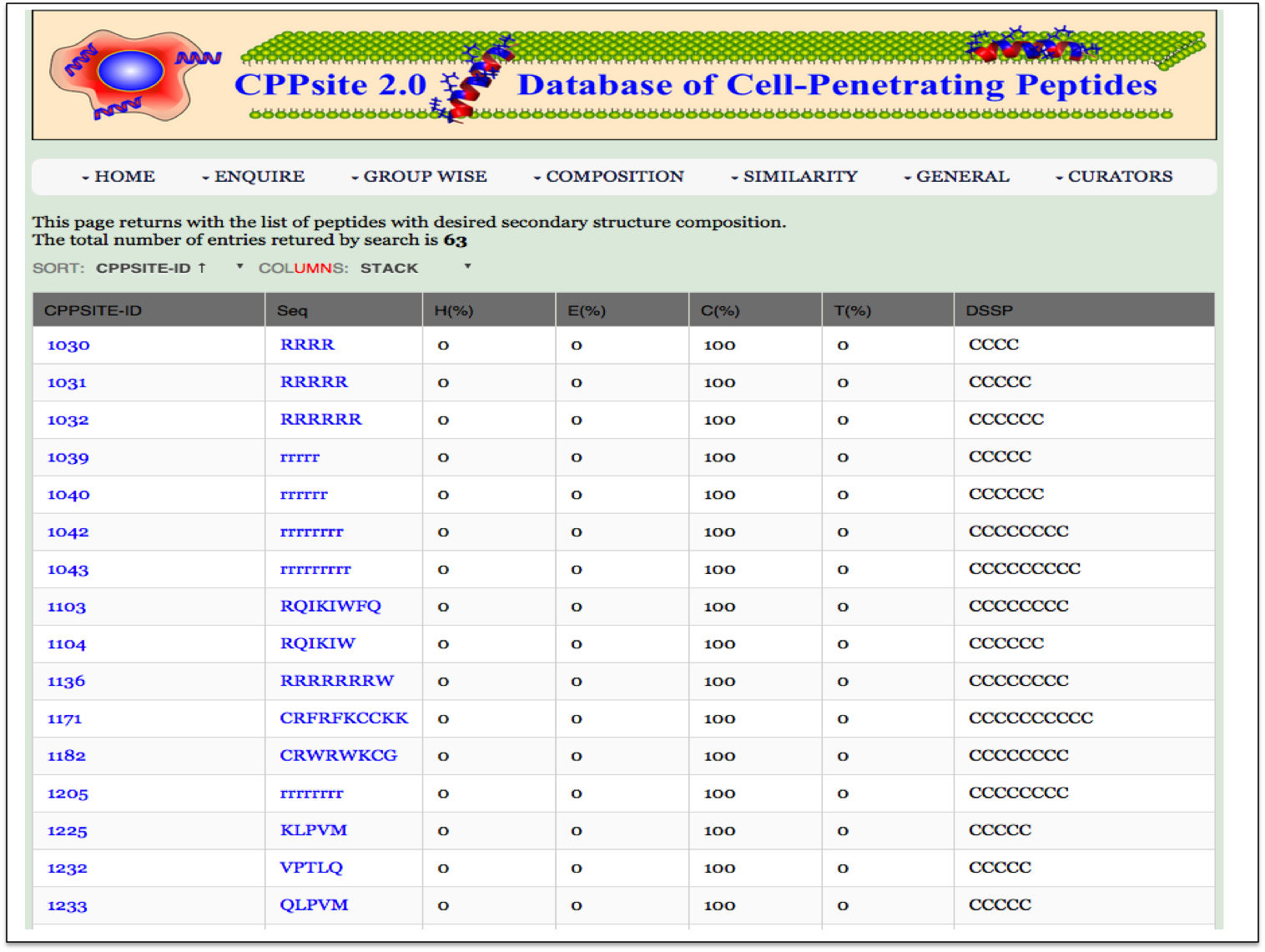 |
Similarity |
BLAST Search Blast-Search page assists users in performing BLAST search against the peptides in CPPsite 2.0. User can submit their peptide with desired BLAST options for performing similarity, server will returns the BLAST output containing list of peptides similar to the query peptide. |
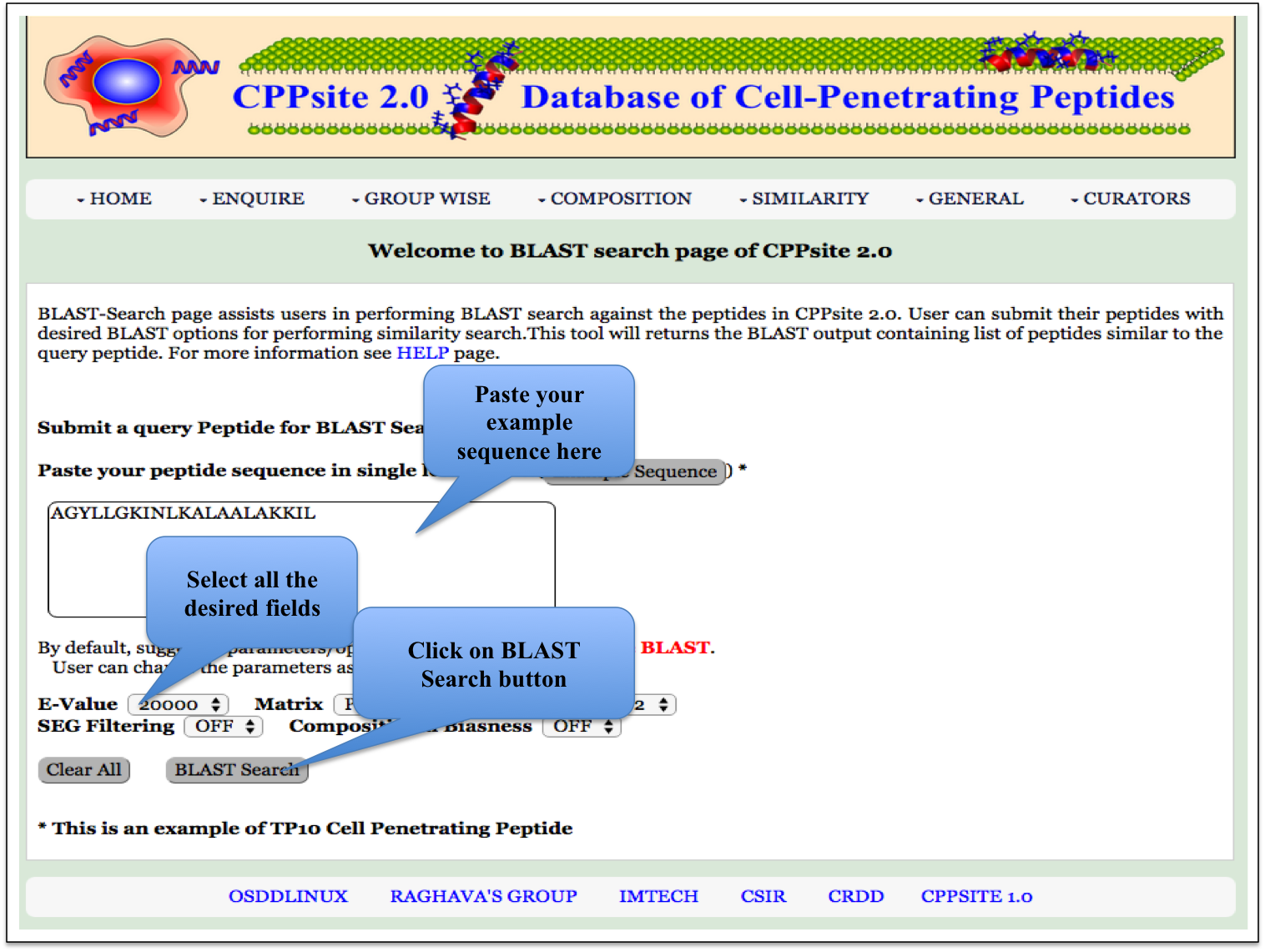 |
| Result |
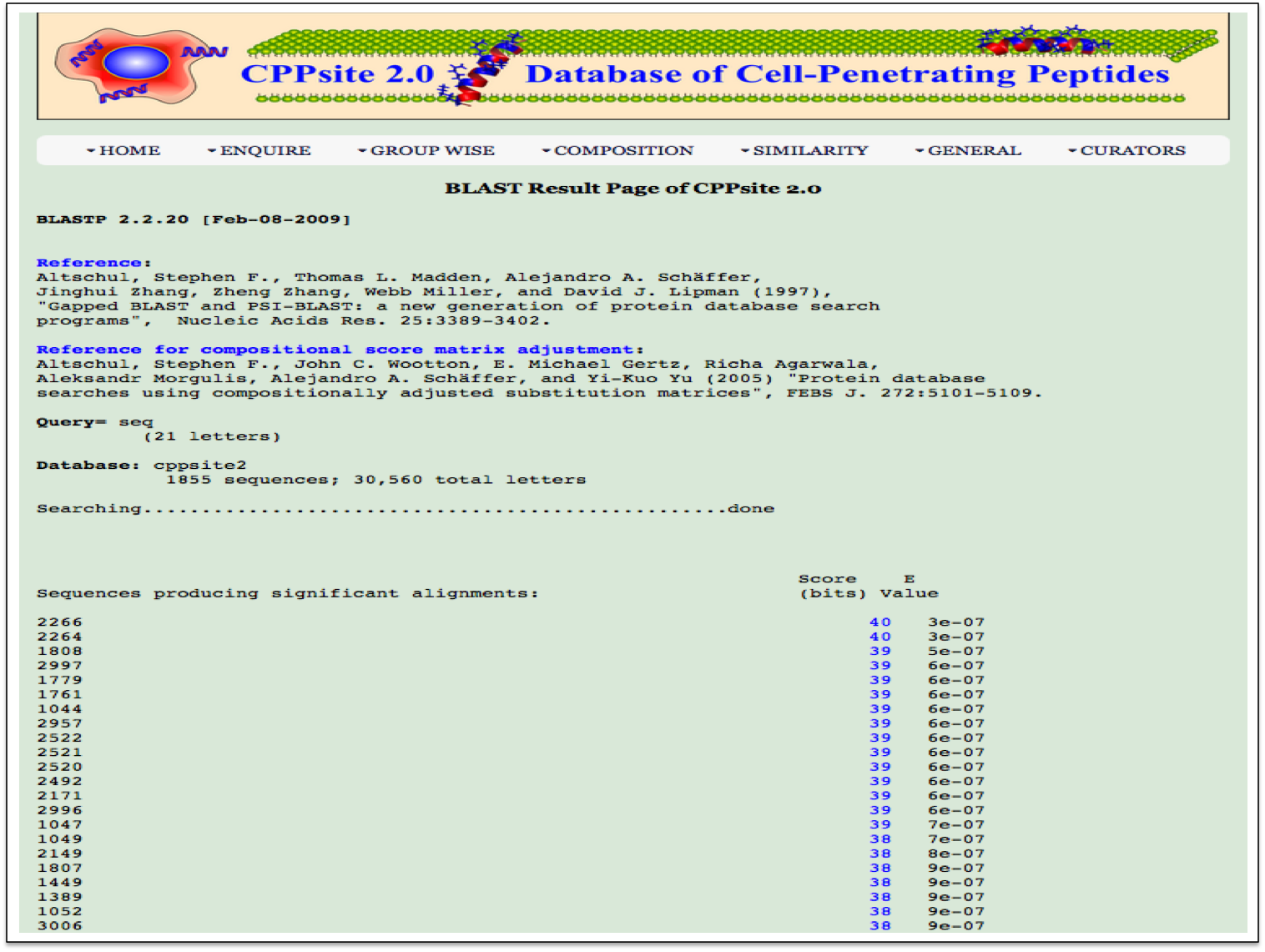 |
2-D Similarity This page provides an option to perform searching of peptides based on secondary structure states of peptides. It will help in searching peptides with similar secondary structure states to the query peptide. Users need to paste their peptide secondary structure in the textbox provided below |
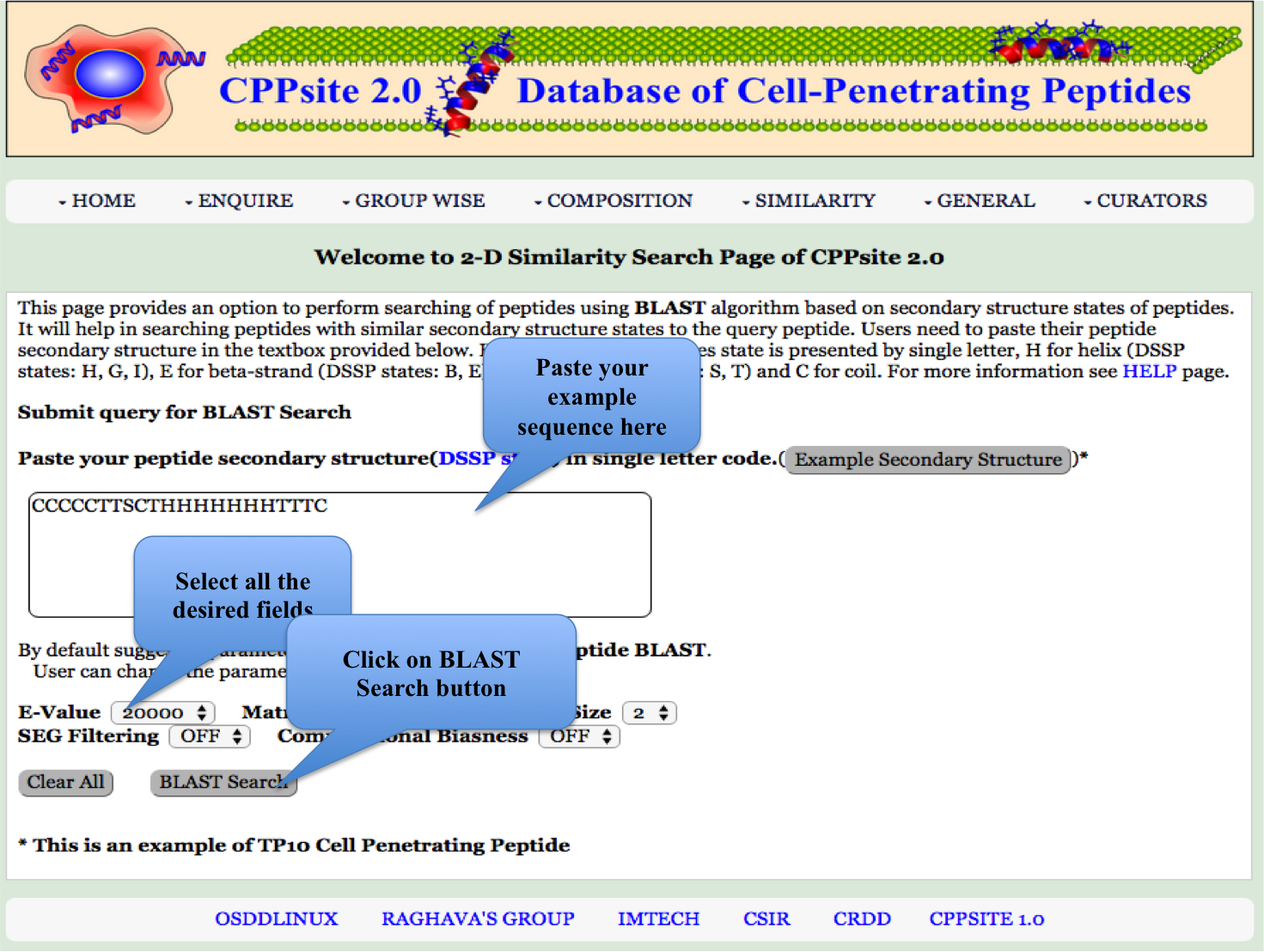 |
| Result |
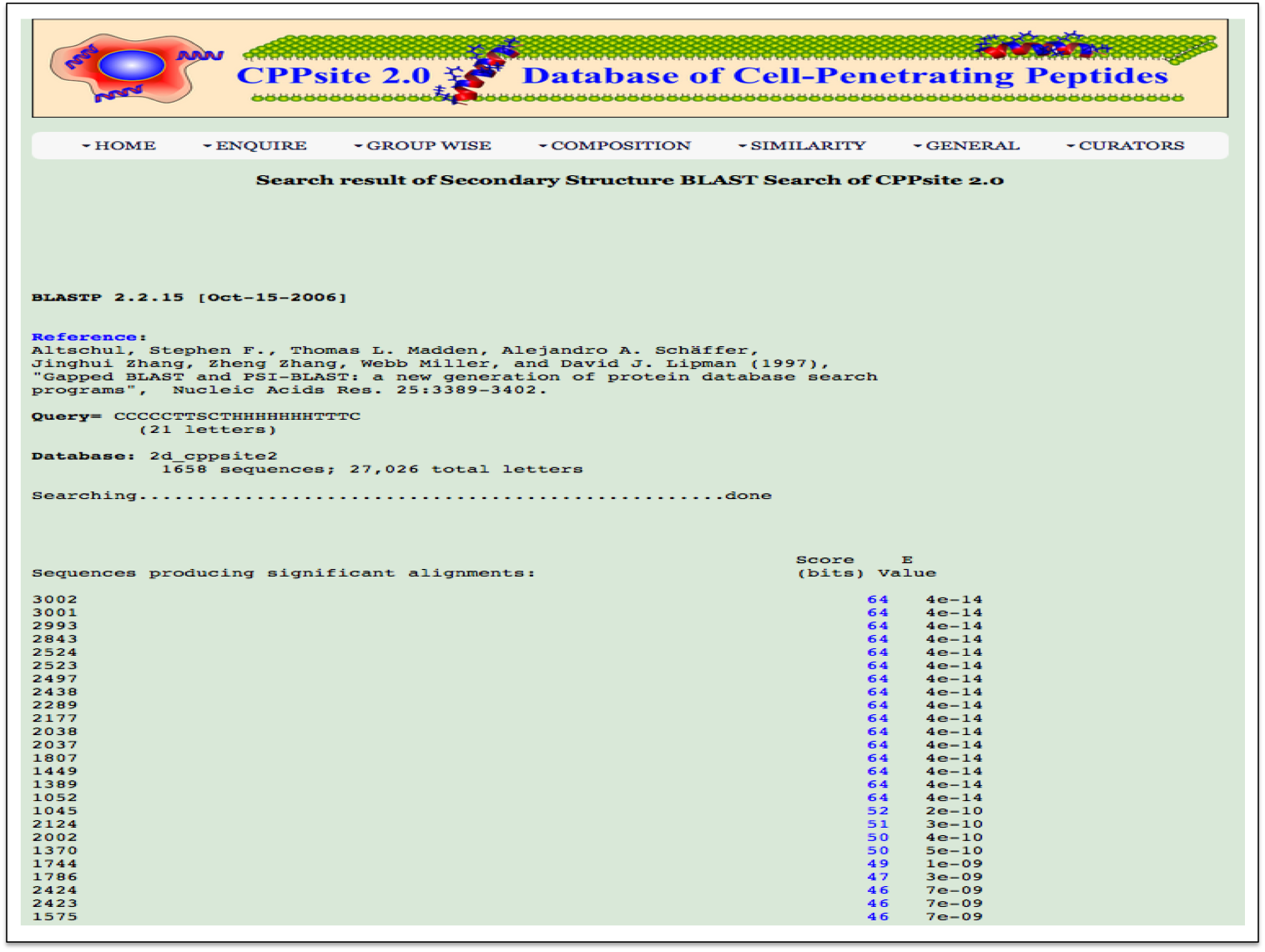 |
Smith-Waterman This page allows users to perform similarity search against peptides in CPPsite 2.0 using Smith-Waterman algorithm. User can submit their peptide; server will returns the list of peptides similar to the query peptide. |
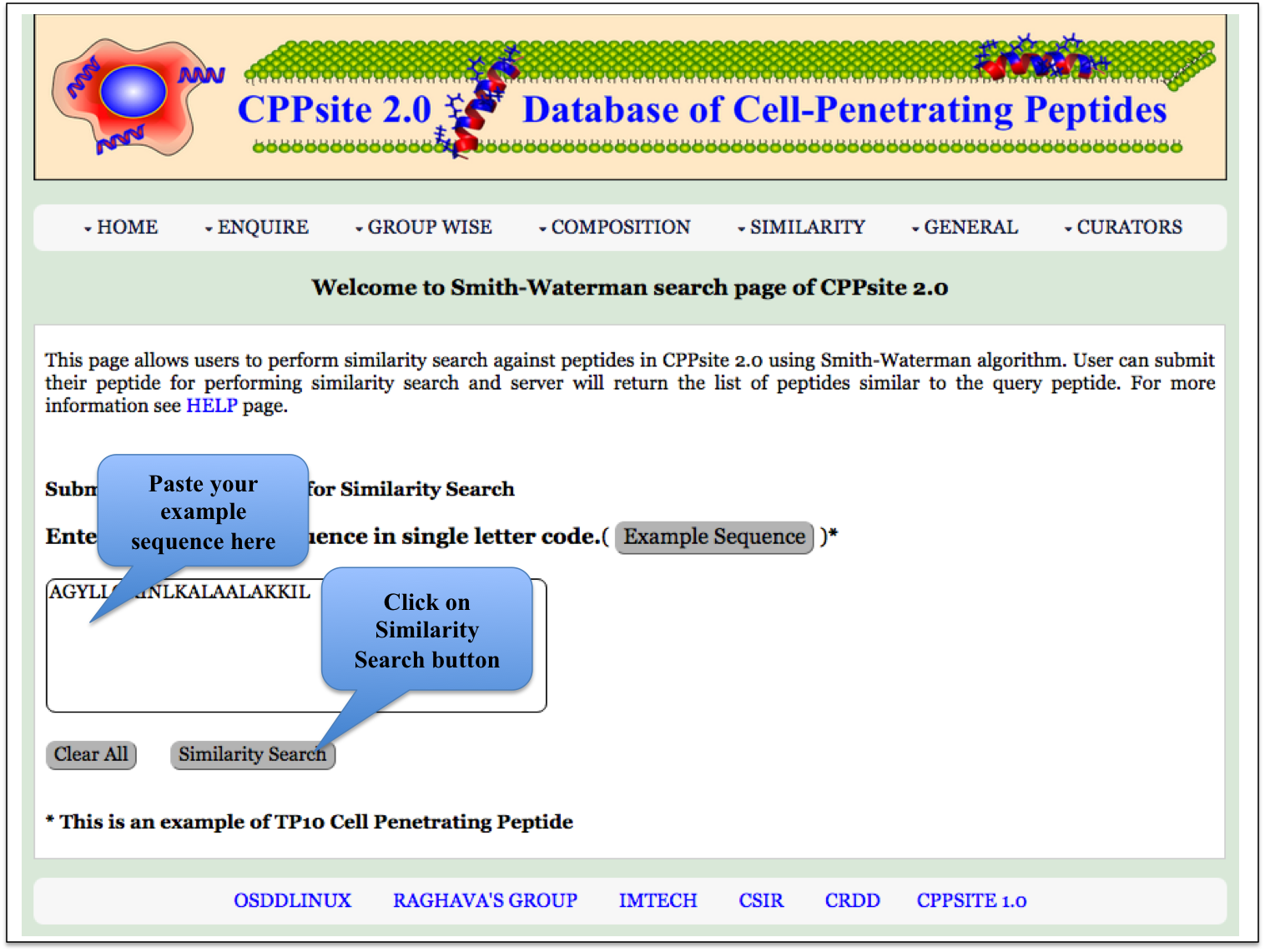 |
| Result |
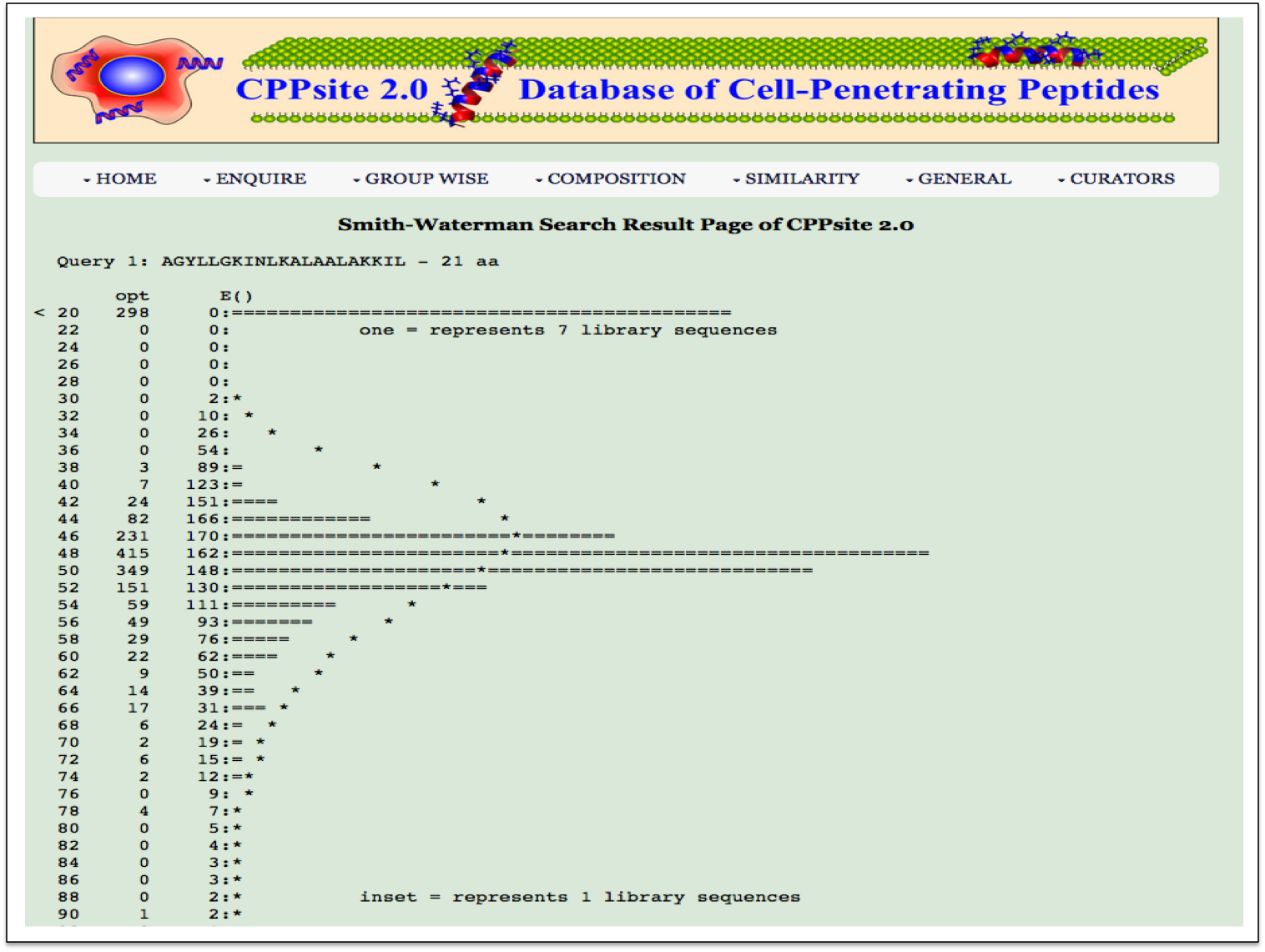 |
3-D Similarity This module had been designed with the aim to provide virtual screening facility to scientific community. Using this tool, users can screen their peptide library and identify similar or exact peptides from various databases. The input for this module could be provided by 1) pasting the peptide structure in defined text area OR 2) Upload the structure file in standard PDB format. |
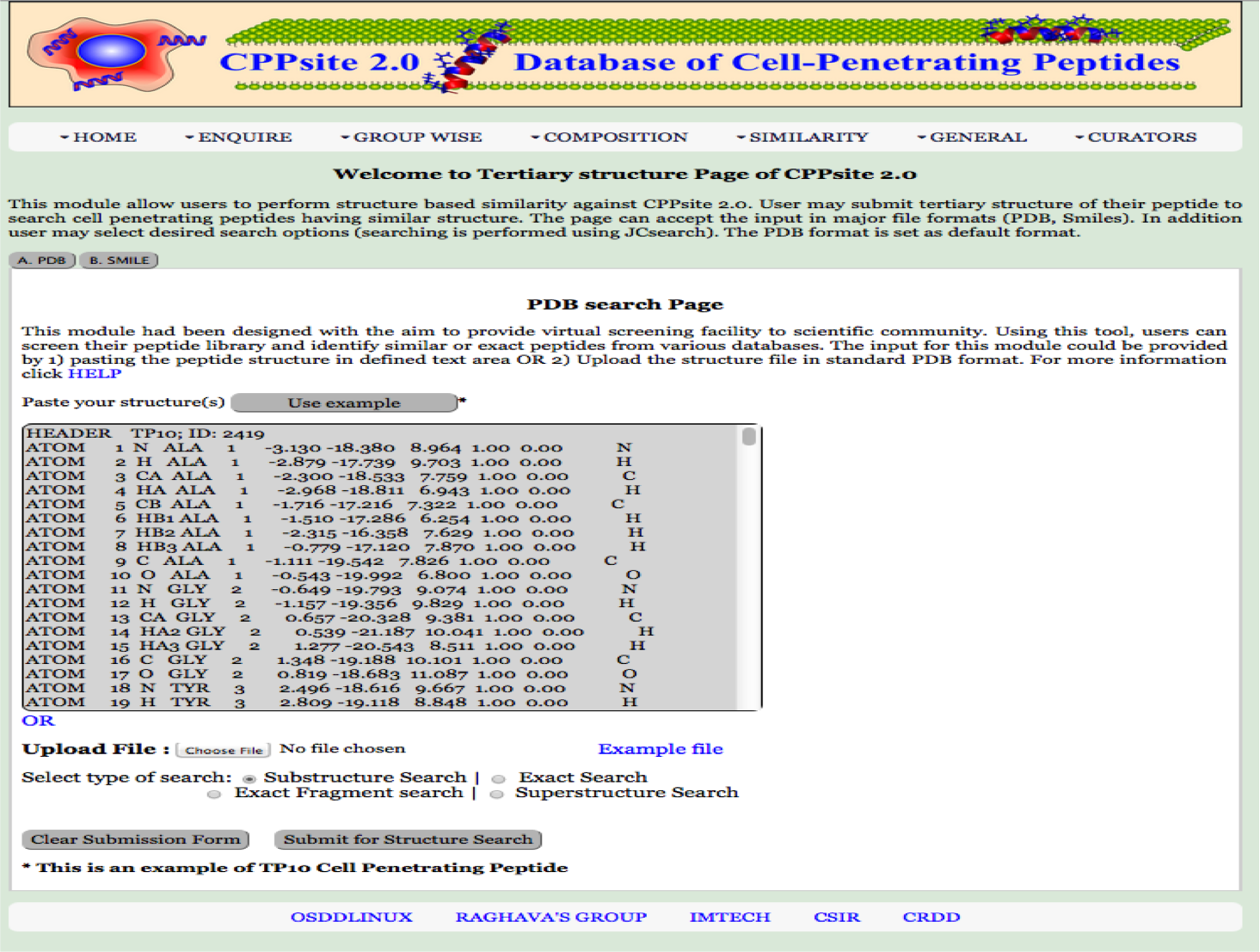 |
| Result |
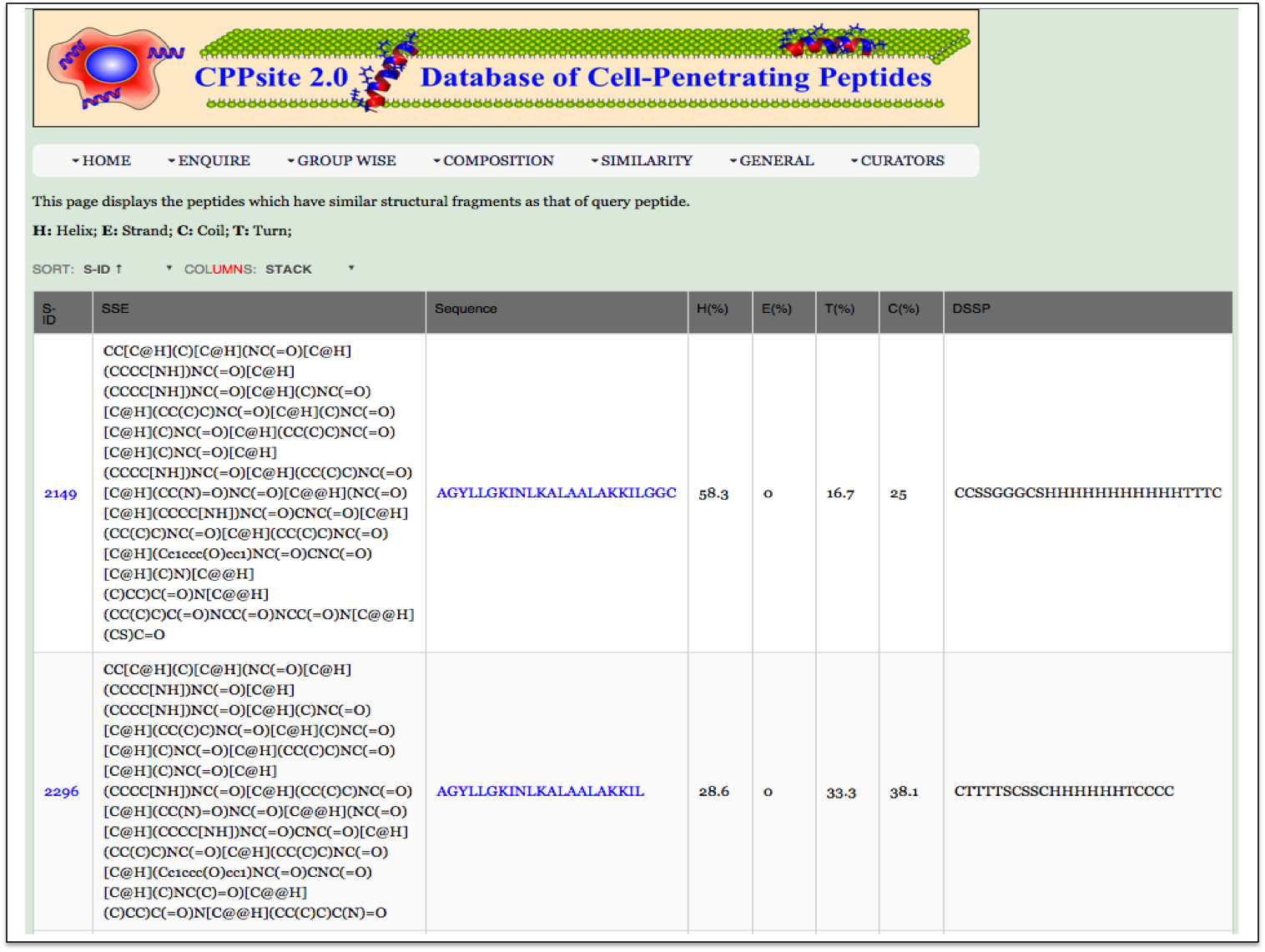 |
Multiple Alignment This page provides the facility to the users to align their sequence with the peptides in CPPsite 2.0. A user inputs multiple fasta sequences in the sequence box and peptide ids of CPPsite-2 in the id box, and gets aligned sequences. |
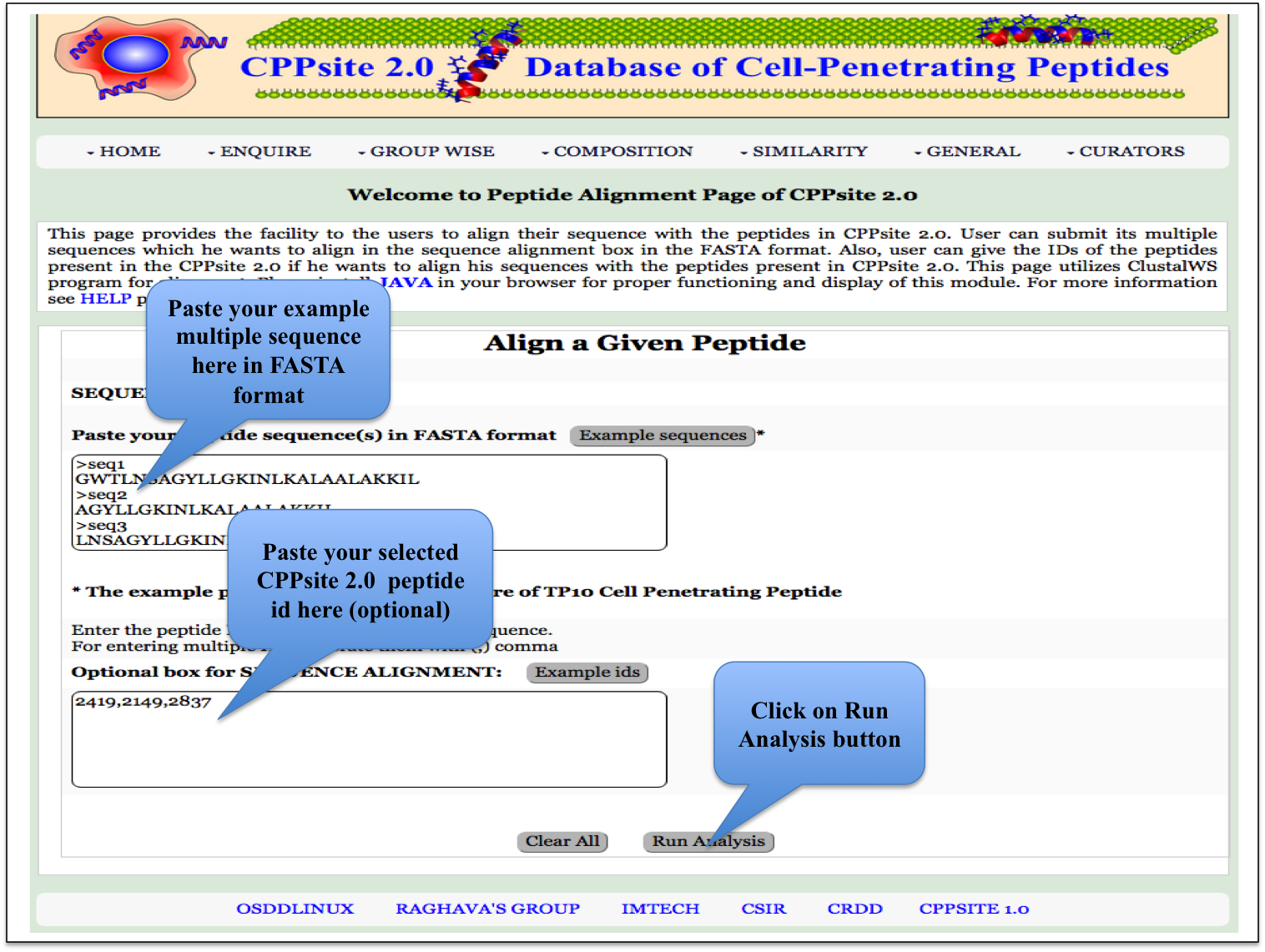 |
| Result |
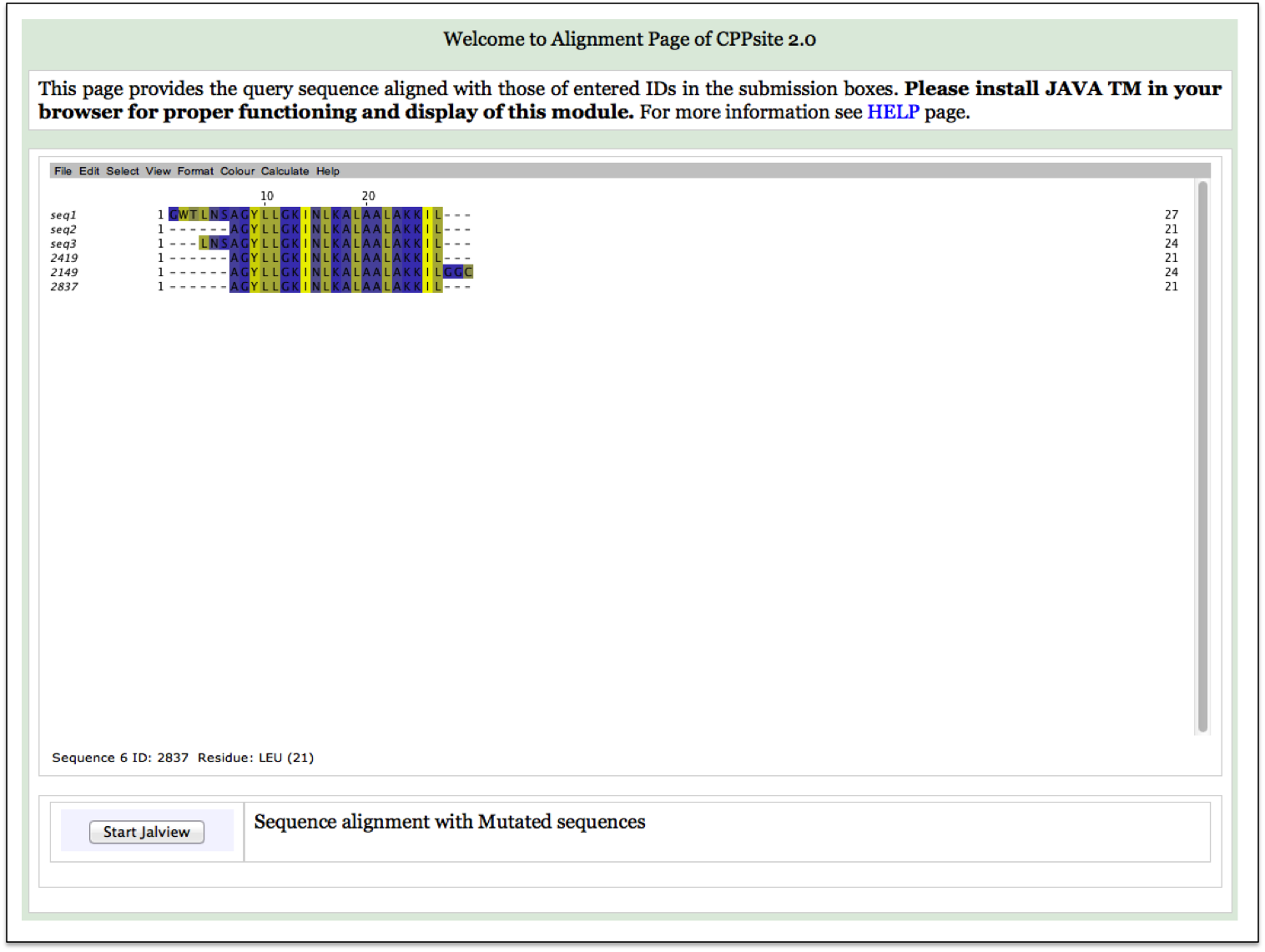 |
Frequently Asked Questions (FAQs) |
|
Q1. What is CPPsite 2.0? Ans .CPPsite 2.0 is updated database of cell penetrating peptides. Q2. Why CPPsite 2.0? Ans. It comprising peptide entries from the literature available on Pubmed and patents. It will give all the information about the cell penetrating peptide. this database will give information about the various modifications done on the CPP and the different types of cargo molecules attached to it for delivery into the cell.This database will helpfull to design novel therapeutic approach. Q3. How to search into CPPsite 2.0? Ans. A user can perform sequence or secondary structure-based searching to identify identical/similar entries present in all the peptide databases integrated in CPPsite 2.0. |