Algorithm of ChAlPred
|
Allergy is the hypersensitivity of the immune system, after exposure to the certain molecules called allergens. There is a wide variety of molecules that can pose a threat as allergens including biological molecules like proteins/peptides and some chemical compounds. The allergy caused by chemical compounds is known as Chemical Allergy. Best of our knowledge no method has been developed for predicting chemicals that can cause allergy. First time an attempt have been made to develop method for predicting allergeinic and non-allergenic chemical compounds.
The prediction server "ChAlPred" has been designed in a very user-friendly manner. Here, on this page, user can get the details of all the algorithms and procedures exploited in the different modules. |
|
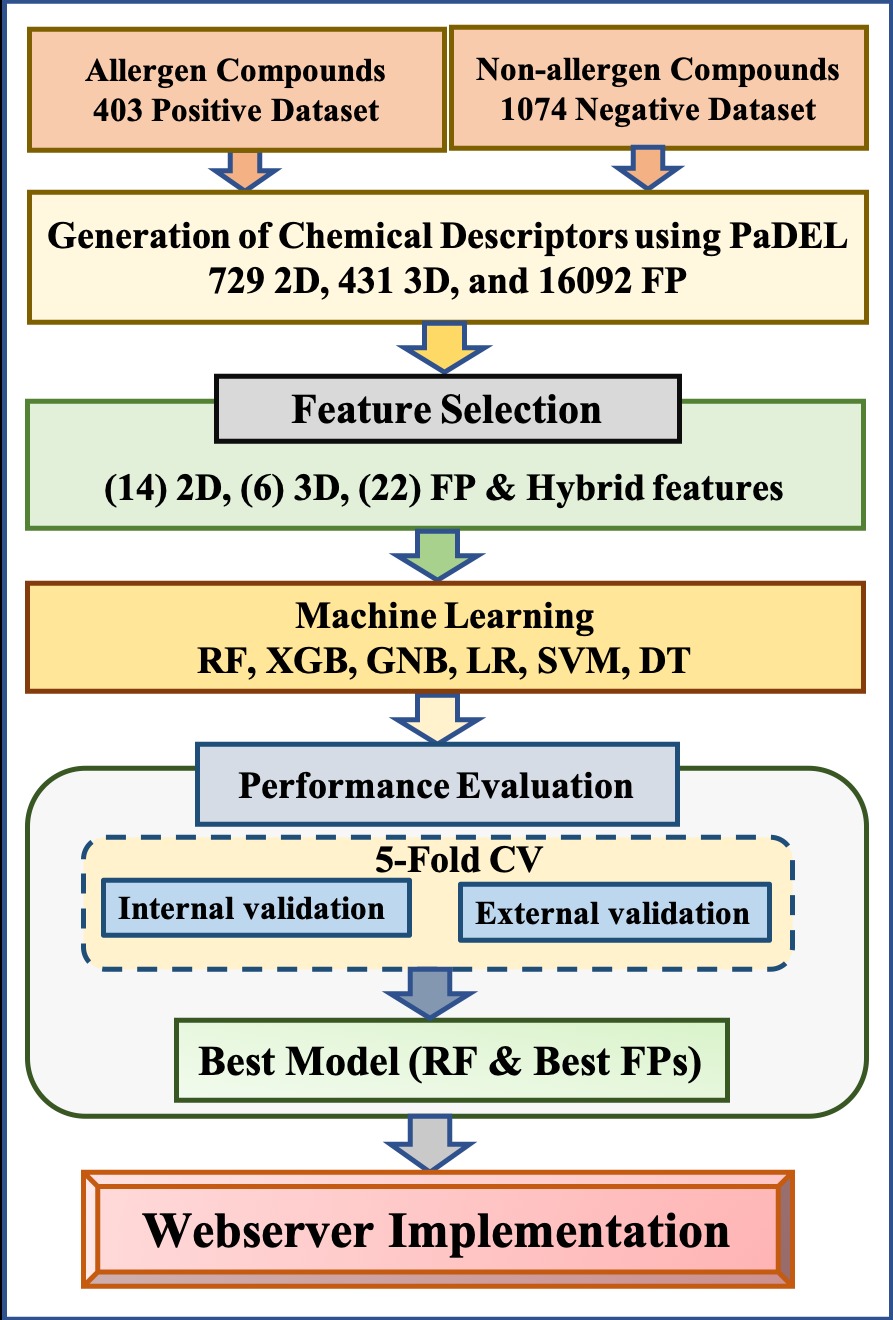
Dataset UsedThe Dataset used in this study is obtained from IEDB. The chemical structures of the compounds were downloaded from ChEBI. The Dataset contains 403 positive data and 1074 negative data. |
This algorithm of the server is relies on the following three models:
Prediction modelThe "Predict" module provides the facility to the user to classify allergenic and non-allergenic chemical compounds. User can provide single or multiple chemical structures in different formats, like SDF, SMILES and MOL. Each molecule would be predicted as allergen and non-allergen based on the selected threshold value. In this study we used various machine learning techniques to develop prediction modules.Draw modelThis module was developed for predicting whether the chemical molecule is allergenic or not. It allows users to draw the chemical structure of the molecule using the Ketcher. Users have the choice to either build a new molecule or edit/modify an existing molecule. To facilitate users, an example structure is provided that can be loaded in the Ketcher by clicking on "Load Example" button. This module has also provided the facility to inform user by the email notification after finishing their job. The outcome of the model will be displayed in tabular form.Analog modelThis module is designed specifically for the users who want to develop analogs of their lead molecule. A user needs to submit a scaffold structure along with the building blocks and linker molecules. Using the submitted information, the web server uses the SmiLib package at the backend and links the building blocks (fragments) to the scaffold using linker to generate all the possible analogs. Each analog is further predicted as allergen and non-allergen, and the results are displayed in tabular form. |
