Welcome to TumorHoPe2.0
TumorHoPe2 is a comprehensive and enhanced version of the original TumorHoPe database, specifically designed to provide detailed information on tumor-homing peptides (THPs). This updated release includes 1103 experimentally validated peptides, and when combined with data from the previous version, offers a total of 1847 entries representing 1297 unique peptide sequences. The database compiles data from two primary sources: phage display libraries—commonly used to identify peptide ligands that bind to tumor-specific markers—and synthetic peptides, which are chemically modified to improve characteristics such as stability, binding affinity, and specificity. Each entry provides key details, including peptide sequences, terminal or chemical modifications, associated cancer cell lines, and targeted tumor types. TumorHoPe2 contains 594 chemically modified peptides, including 255 with N-terminal and 195 with C-terminal modifications. These peptides have been tested on 172 cancer cell lines, targeting 37 distinct tumor types.
Statistcs
Total Peptides
1847

Unique Peptides
1297

Tumor Count
37
Cell Line
172
About TumorHoPe2.0
 Importance of Peptides
Importance of Peptides

Total Peptides
1847

Unique Peptides
1297

Tumor Count
37

Cell Line
172

 Importance of Peptides
Importance of Peptides
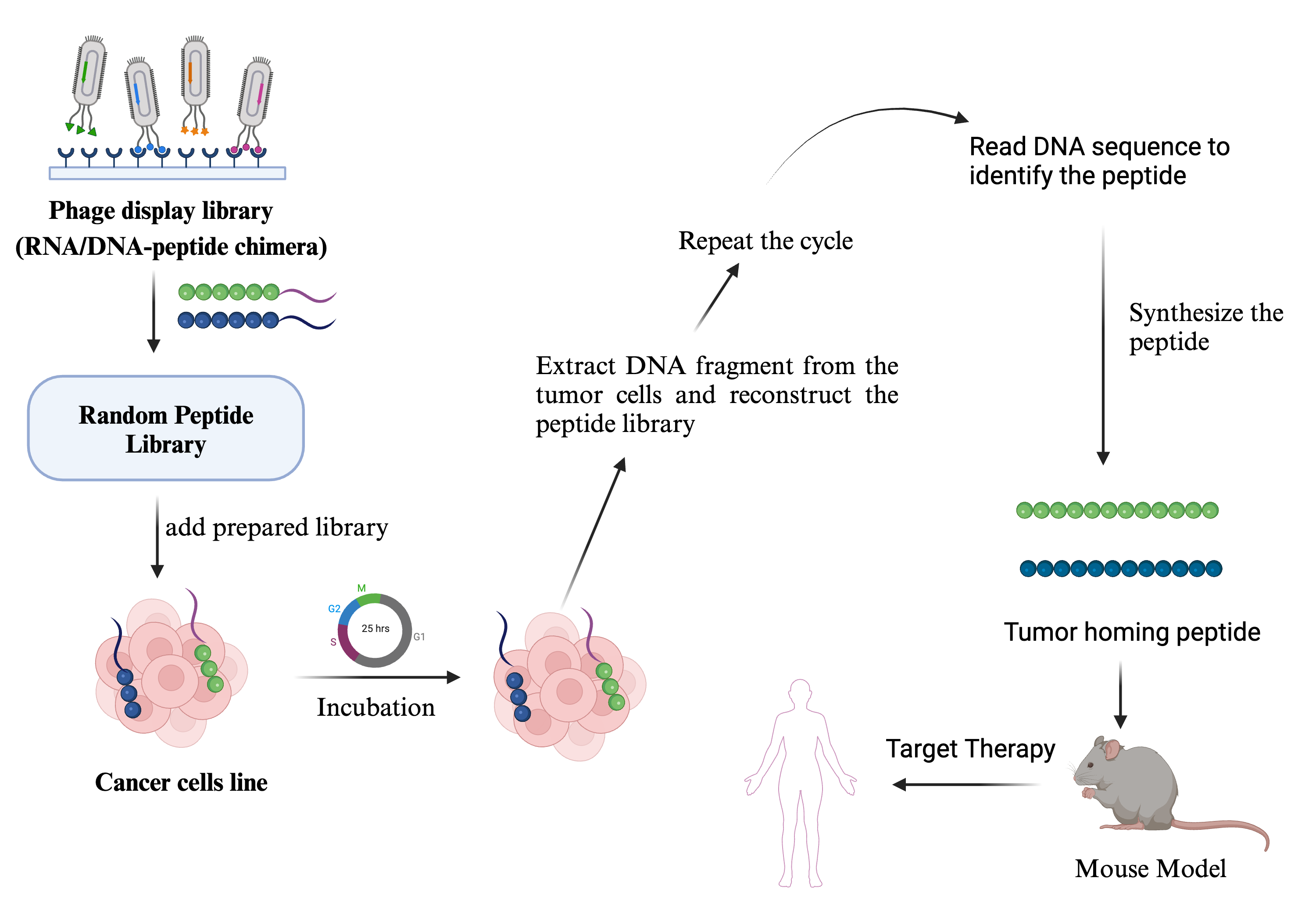
Poor selectivity of chemotherapeutic drugs for cancer is a major challenge for successful clinical outcomes. Conjugation of a drug with a homing peptide may enhance the selectivity and efficacy of the therapy. Current efforts are focused on tumor homing peptides that may target tumor tissues.
 Information about Peptides
Information about Peptides
Tumor Homing Peptide Database has been developed using extensive literature search. It contains detailed information about tumor-targeting/homing peptides. Each entry includes information about a peptide's sequence, source, target tumor, target cell, biomarker, applications, and clones. Experimental details like phage display libraries used, cell lines, in vivo animal models, and payloads used are also incorporated.
 Sequence Analysis of Peptides
Sequence Analysis of Peptides
We have computed amino acid composition, amino acid frequency, and other physical properties of each peptide. Users can search peptides with desired amino acid frequency or compositions or physico-chemical properties.
 Structure of Peptides
Structure of Peptides
In order to design a peptide, it is important to understand its tertiary structure. To provide structural information, we have predicted the tertiary structure of tumor homing peptides using PEPstr. Secondary structures of these peptides were assigned using the program DSSP.
 Important Tools
Important Tools
This resource is equipped with several user-friendly tools, including but not limited to:
- Keyword Search for searching any field
- Peptide Search for searching peptide sequences
- BLAST Search for similarity searches