
- Home
- Predict
- Draw
- Analog Design
- Datasets
General ▾
|
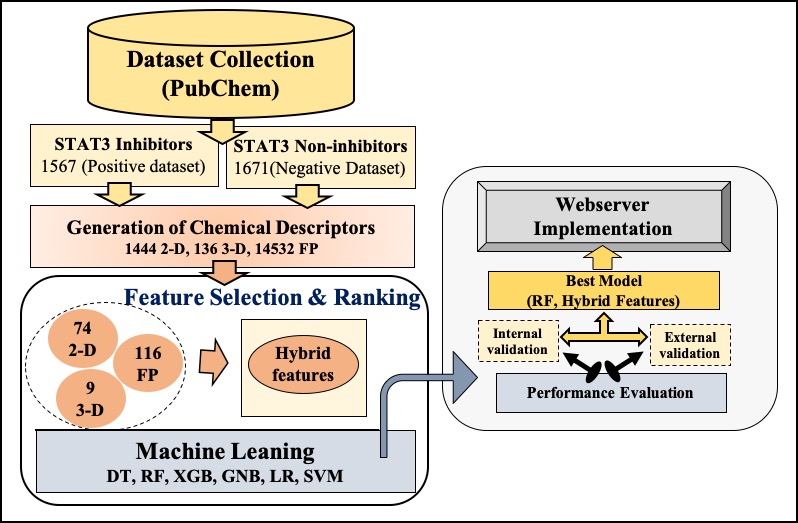
IL6 mediated activation of STAT3 is responsible to proliferate proinflammatory responses that leads to promotion of cytokine storm. Thus, STAT3 inhibitors may play a crucial role in managing pathogenesis of COVID-19. This paper describes a method developed for predicting inhibitors against the IL6-mediated STAT3 signaling pathway.
The prediction server for STAT3 inhibitor molecules prediction has been designed in a very user-friendly manner. Here, on this page, user can get the details of all the algorithms and procedures exploited in the different modules. |
|
Dataset UsedPositive Dataset: STAT3 inhibitors chemical compoundsNegative dataset: STAT3 non-inhibitors chemical compounds |
This algorithm of the server is relies on the following three models:
Prediction modelThis module has been developed to predict the chemical molecules as Signal Transducer and Activator of Transcription 3 (STAT3) inhibitor or non-inhibitor. Here the users are allowed to paste or upload a file with multiple molecules in different file formats like; SMILES, SDF, and MOL format, and each molecule would be predicted as inhibitor or non-inhibtor of STAT3 based on the selected threshold value.Draw modelThis module was developed for predicting whether the chemical molecule is an inhibitor of STAT3 or not. It allows users to draw the chemical structure of the molecule using the Ketcher. Users have the choice to either build a new molecule or edit/modify an existing molecule. To facilitate users, an example structure is provided that can be loaded in the Ketcher by clicking on "Load Example" button. This module has also provided the facility to inform user by the email notification after finishing their job. The outcome of the model will be displayed in tabular form.Analog Design modelThis module is designed specifically for the users who want to develop analogs of their lead molecule. A user needs to submit a scaffold structure along with the building blocks and linker molecules. Using the submitted information, the web server uses the SmiLib package at the backend and links the building blocks (fragments) to the scaffold using linker to generate all the possible analogs. Each analog is further predicted as STAT3 inhibitor and non-inhibitor, and the results are displayed in tabular form. |