Welcome to Help Page of SAPdb
This page provides help to the users of SAPdb database on how to use various modules that are integrated in SAPdb. For each module which is implemented in SAPdb, here we provide the figures and description on how to use that module.
Search Module
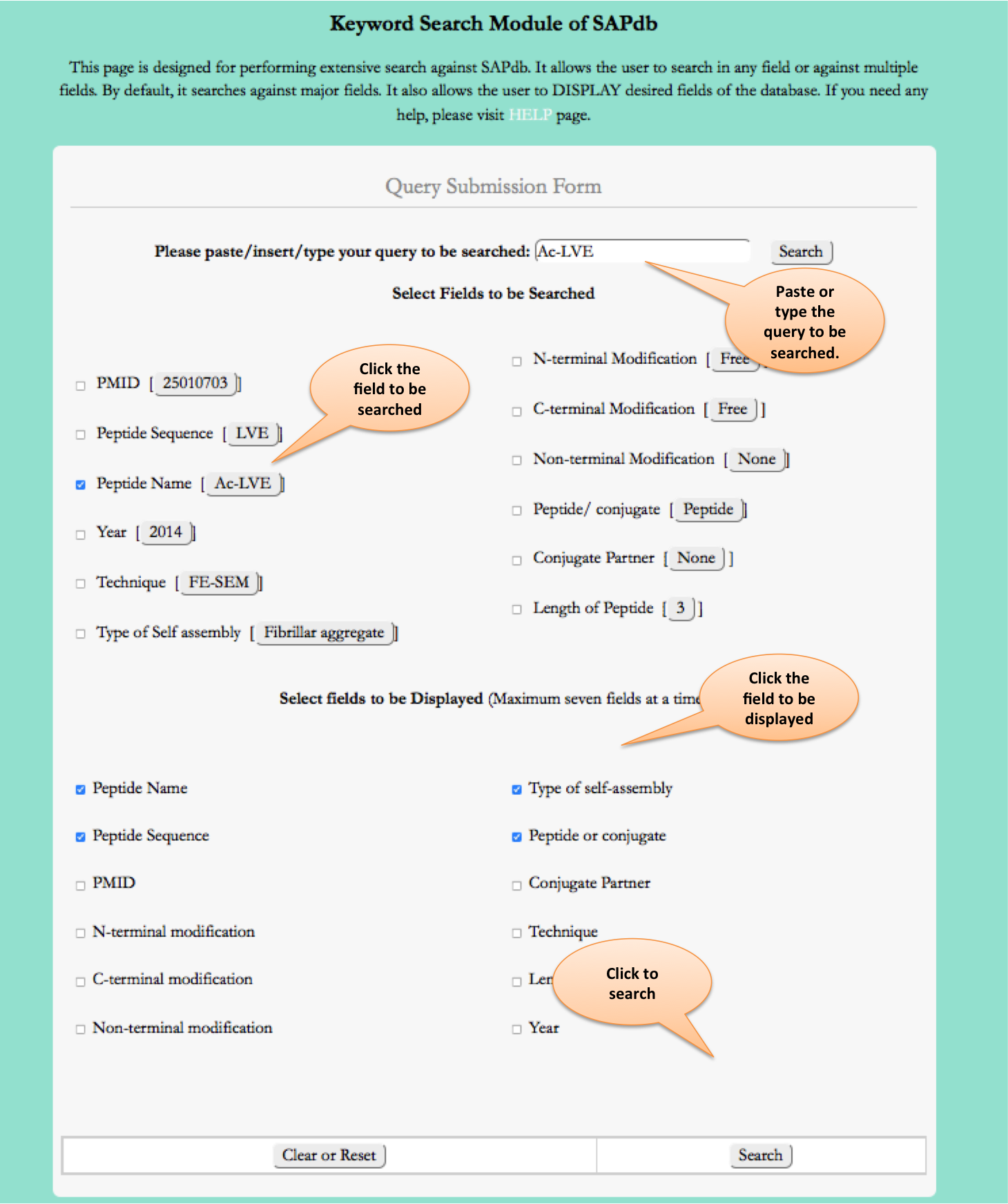
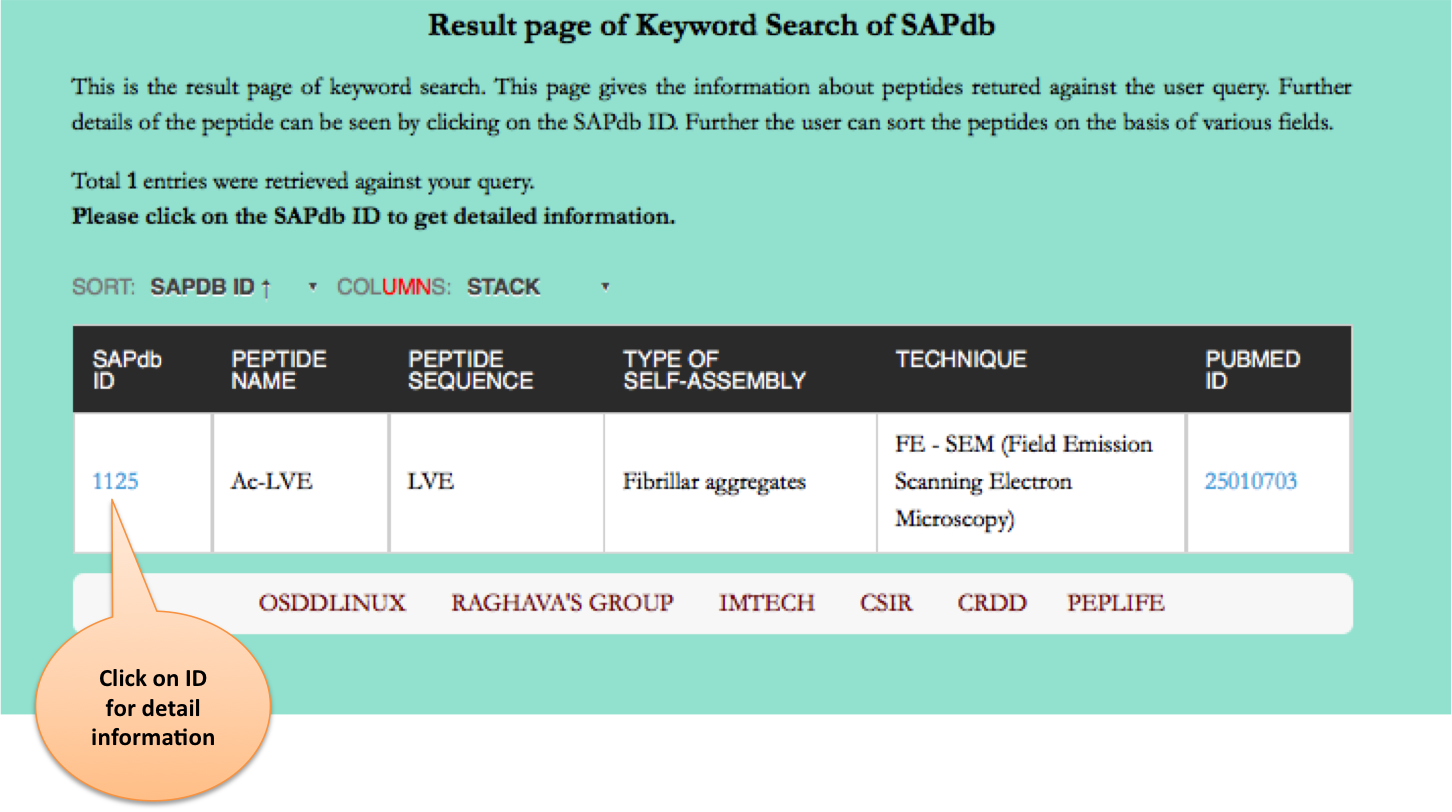
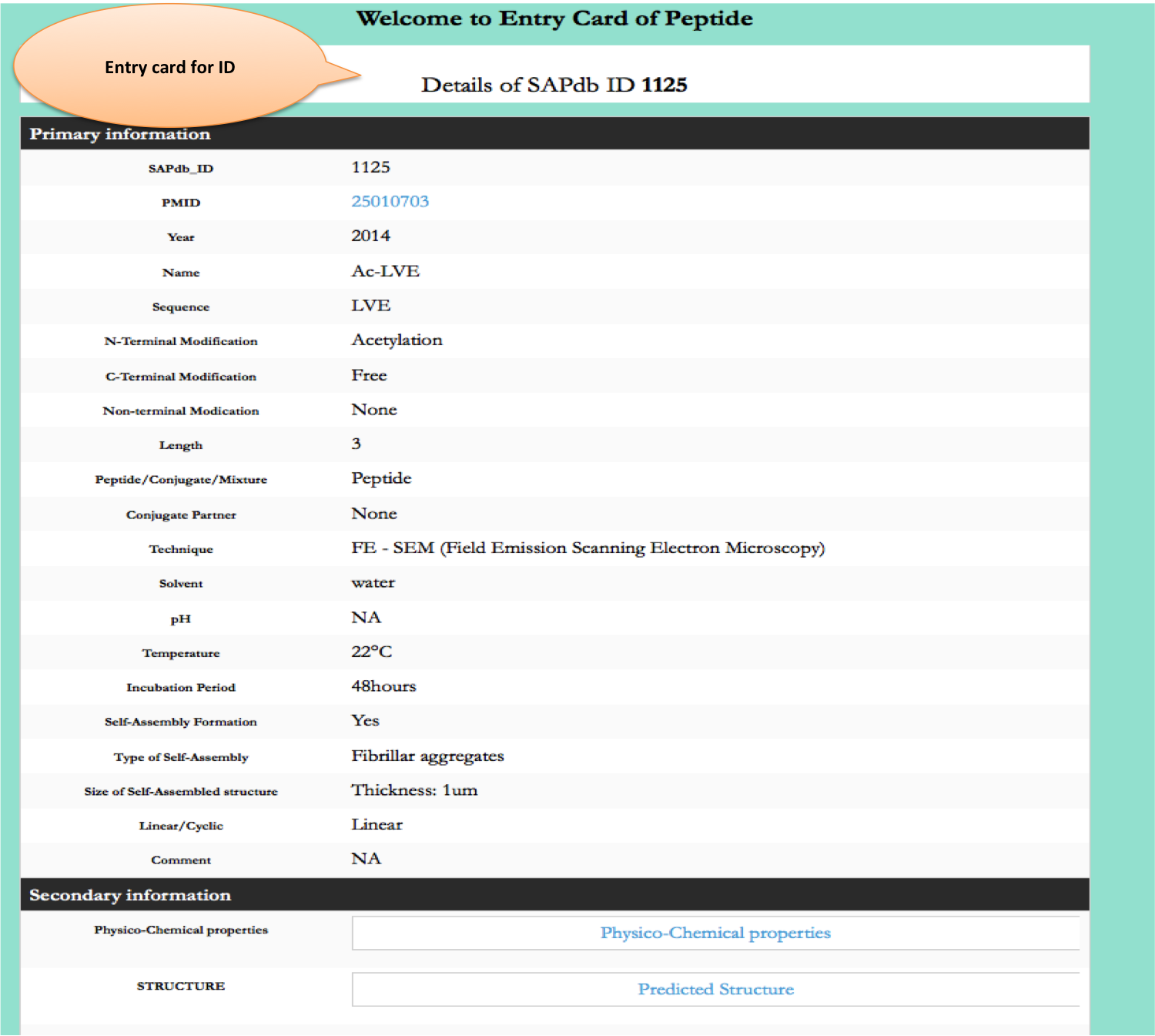
Keyword Search
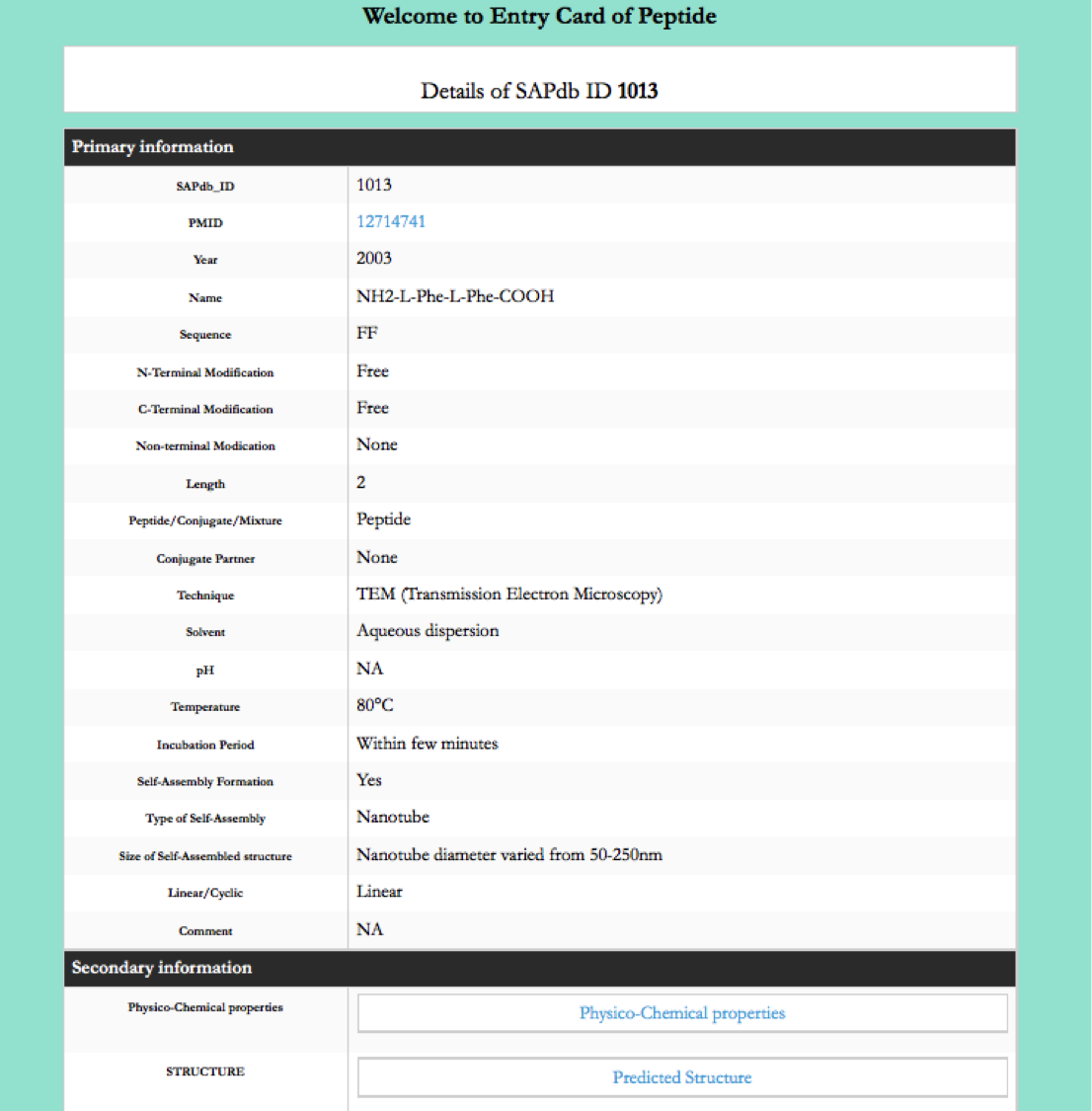
This module displays the simple search page of SAPdb. Clicking on the required field will allow the user to browse the specific information in the database.
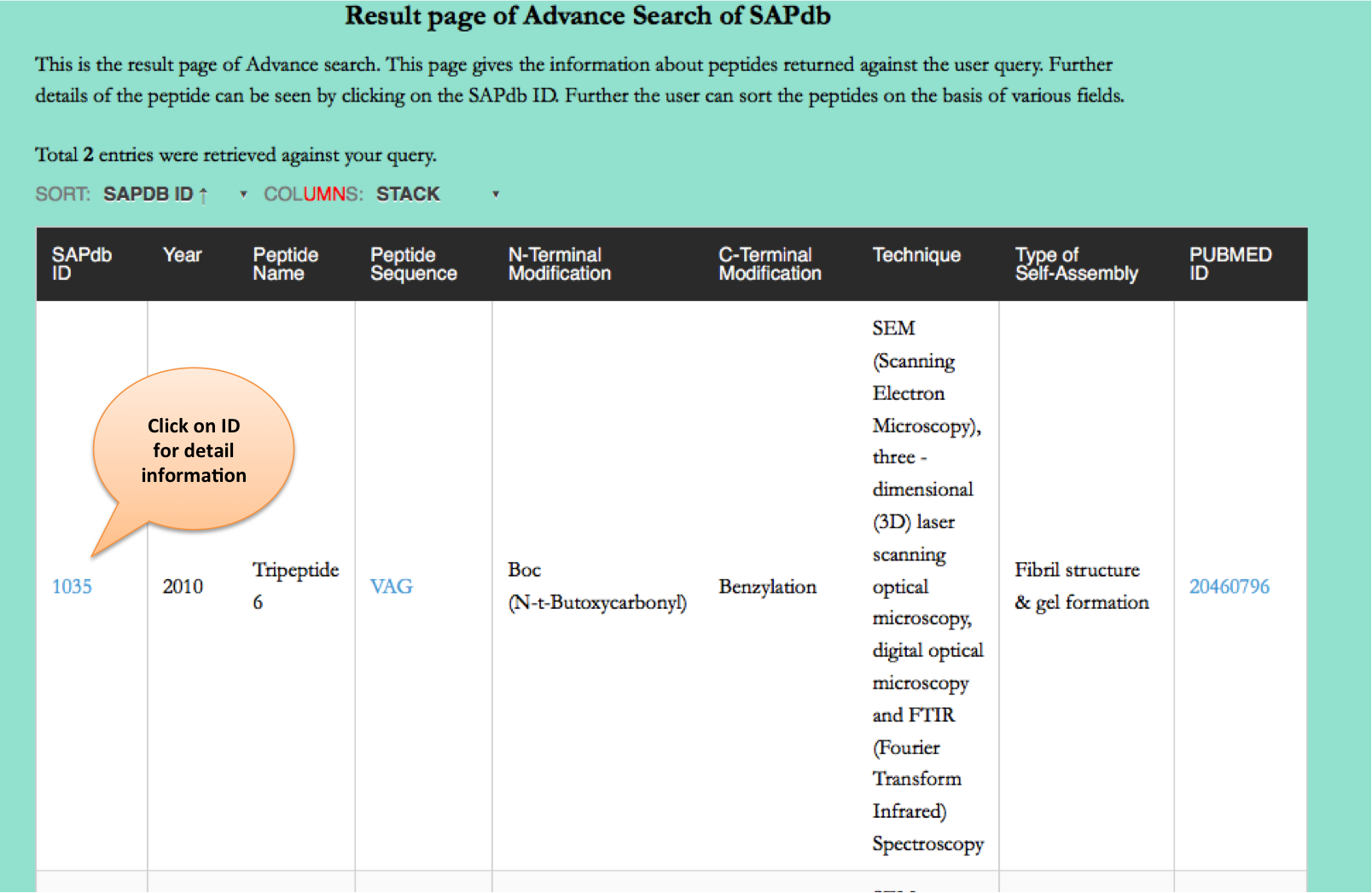
Advance Search
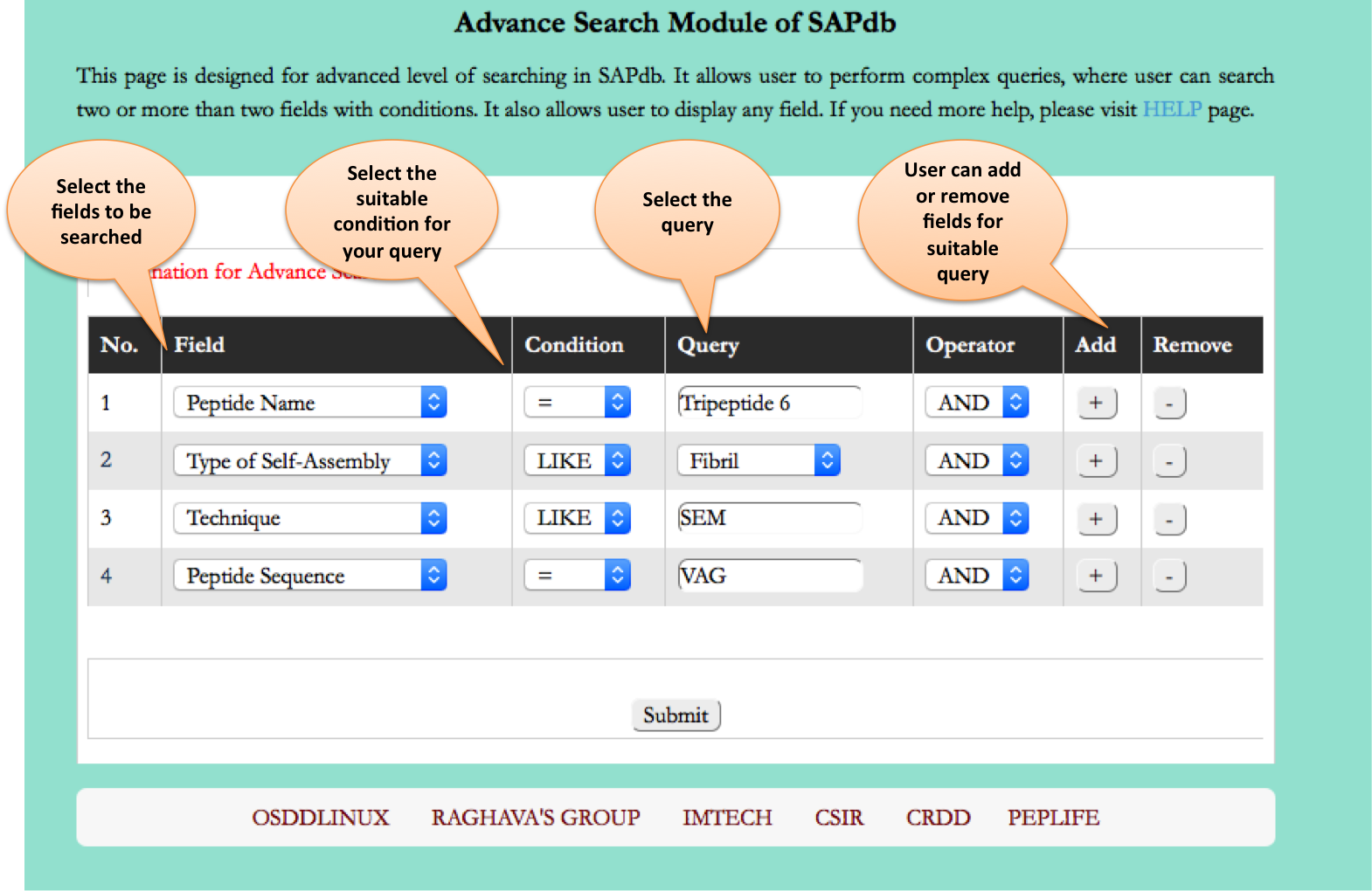
This page provides the advanced search facility in SAPdb. User can browse the page by selecting two or more than two fileds.

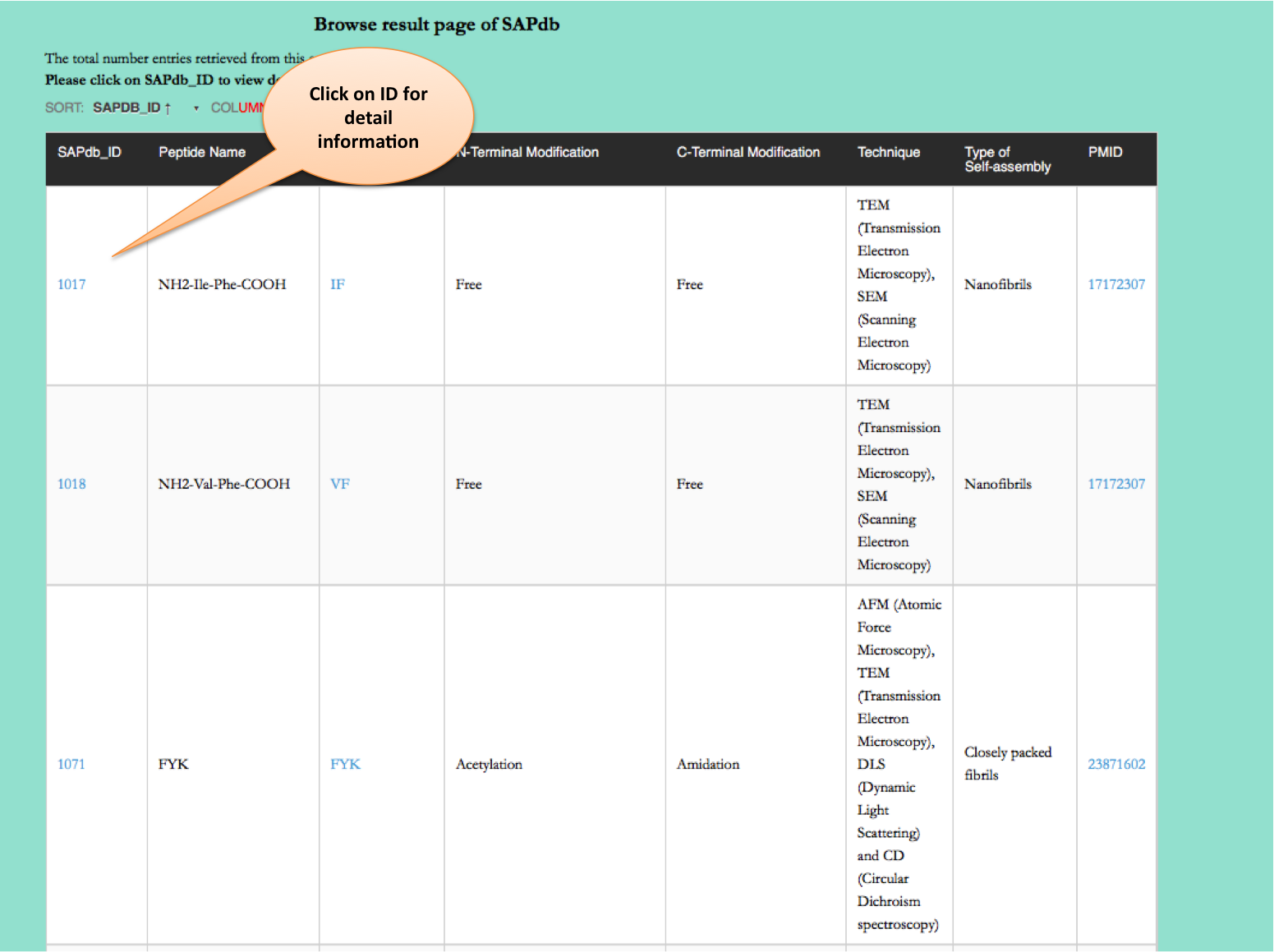
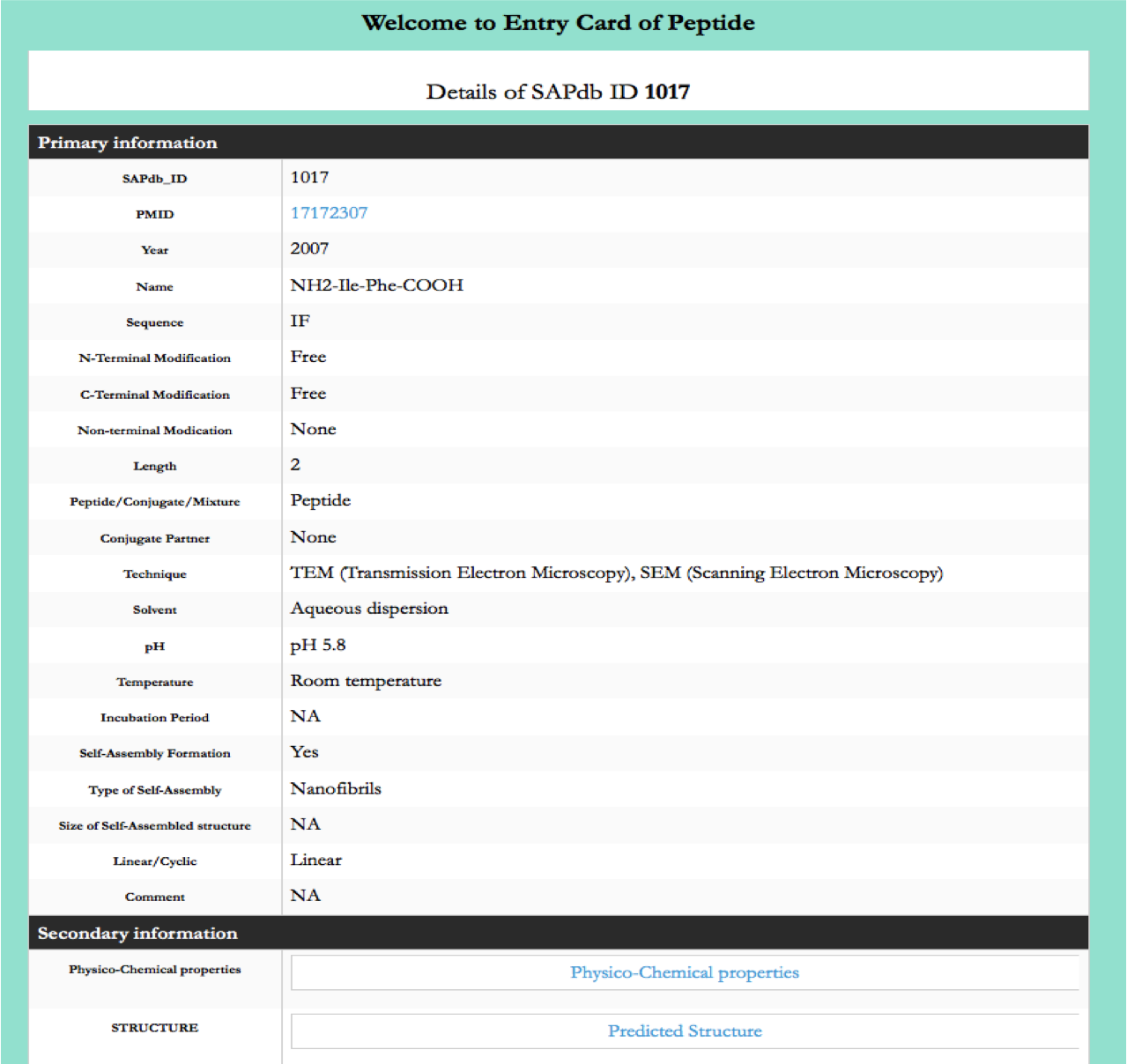
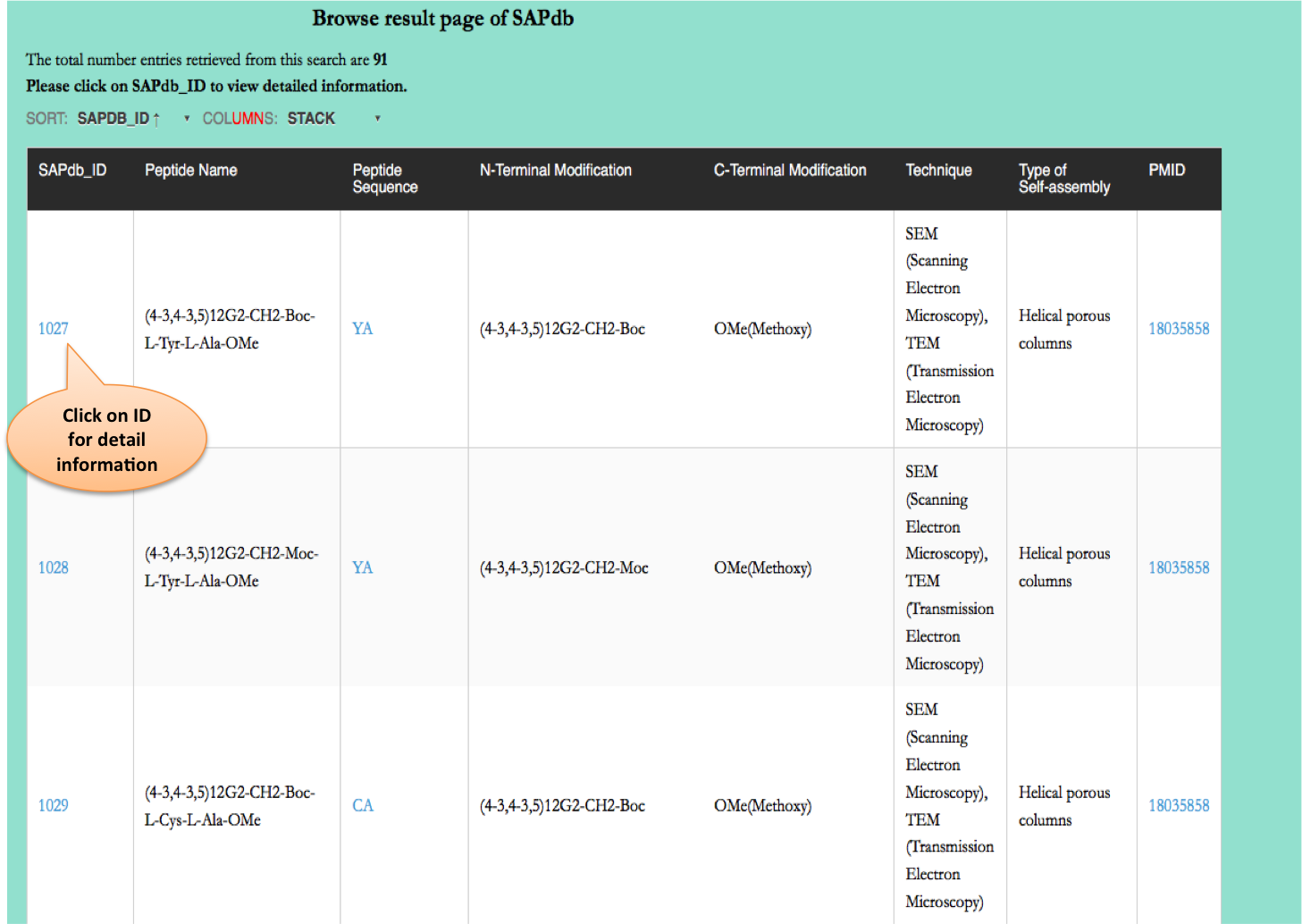
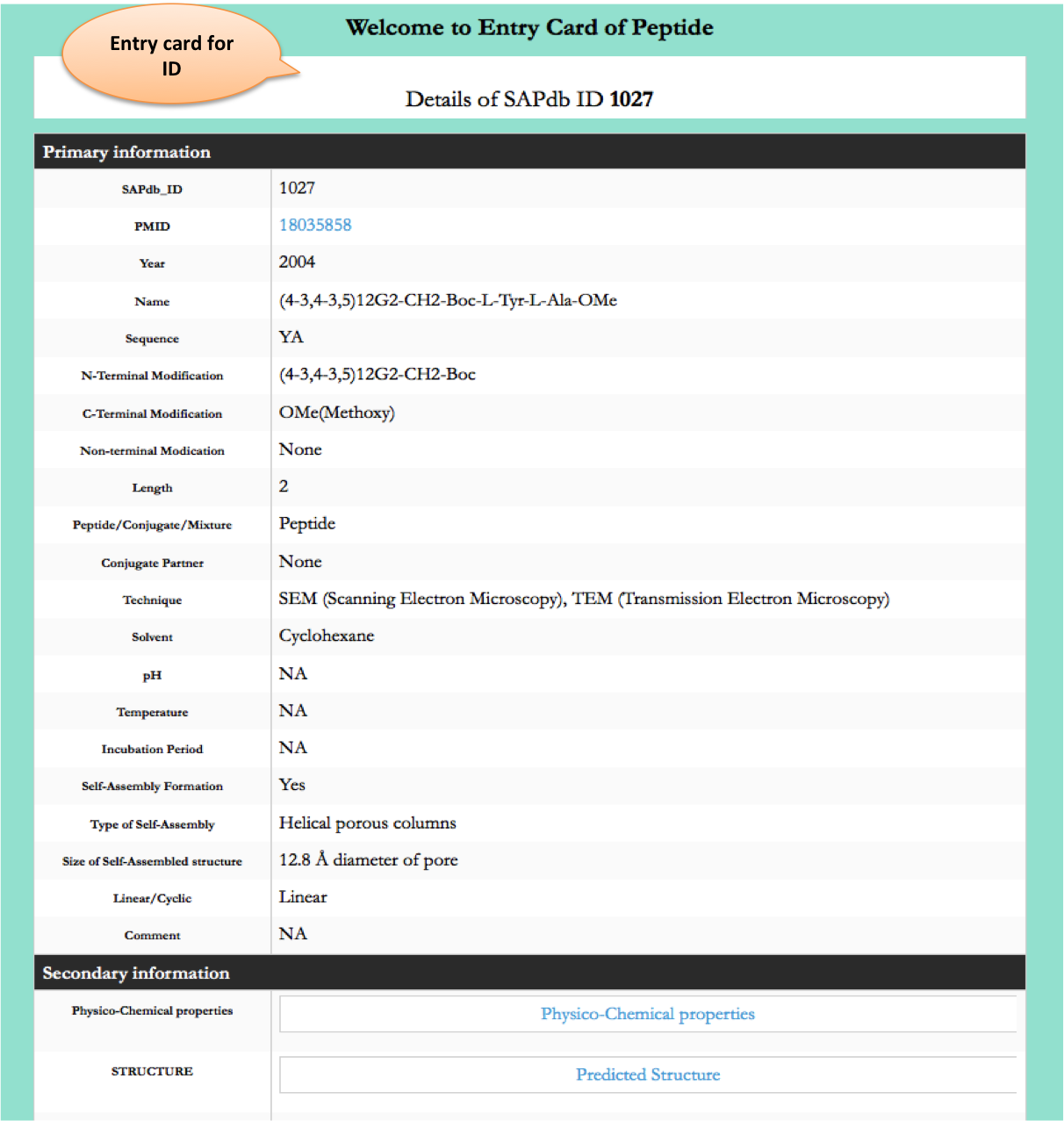
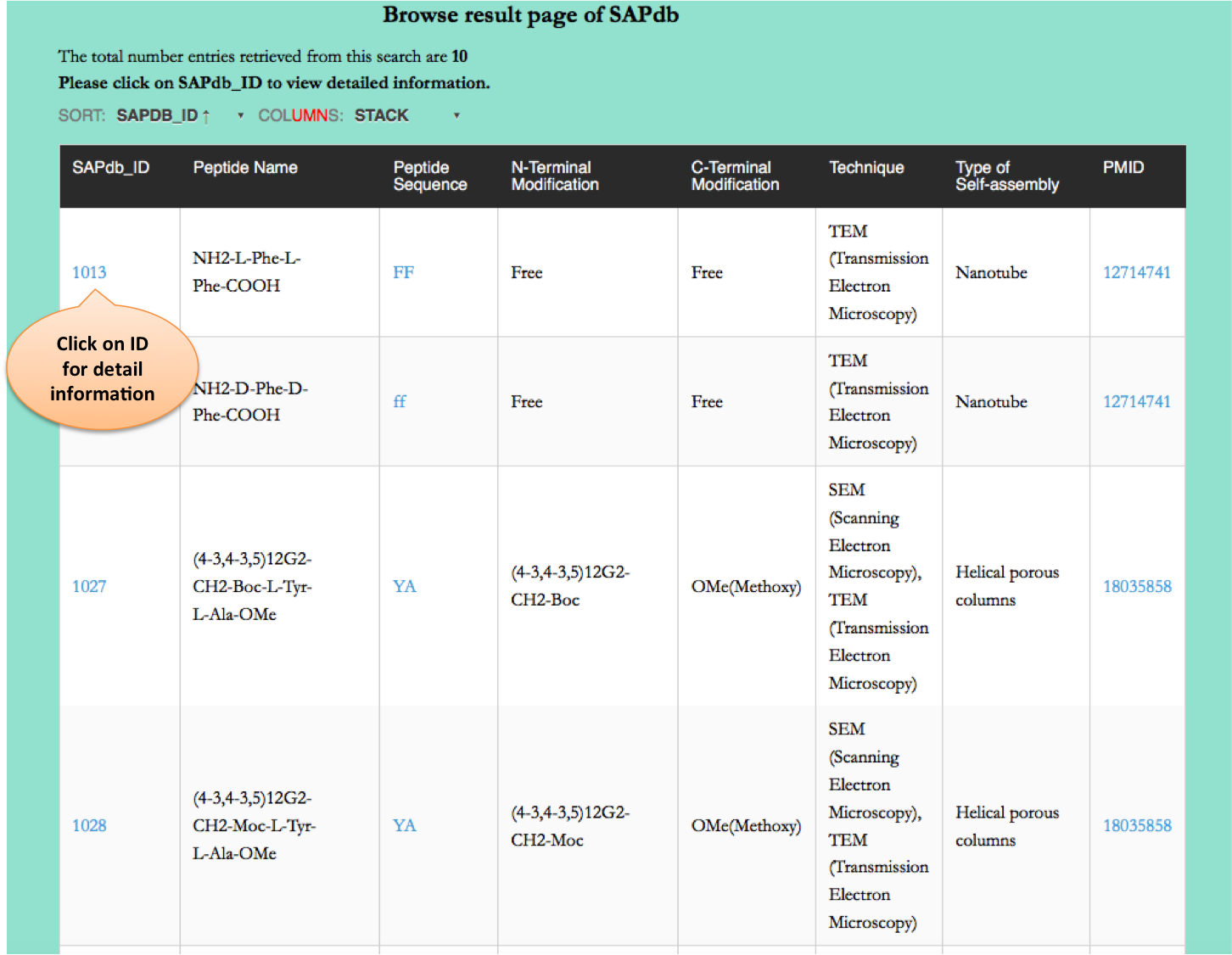
Browse page
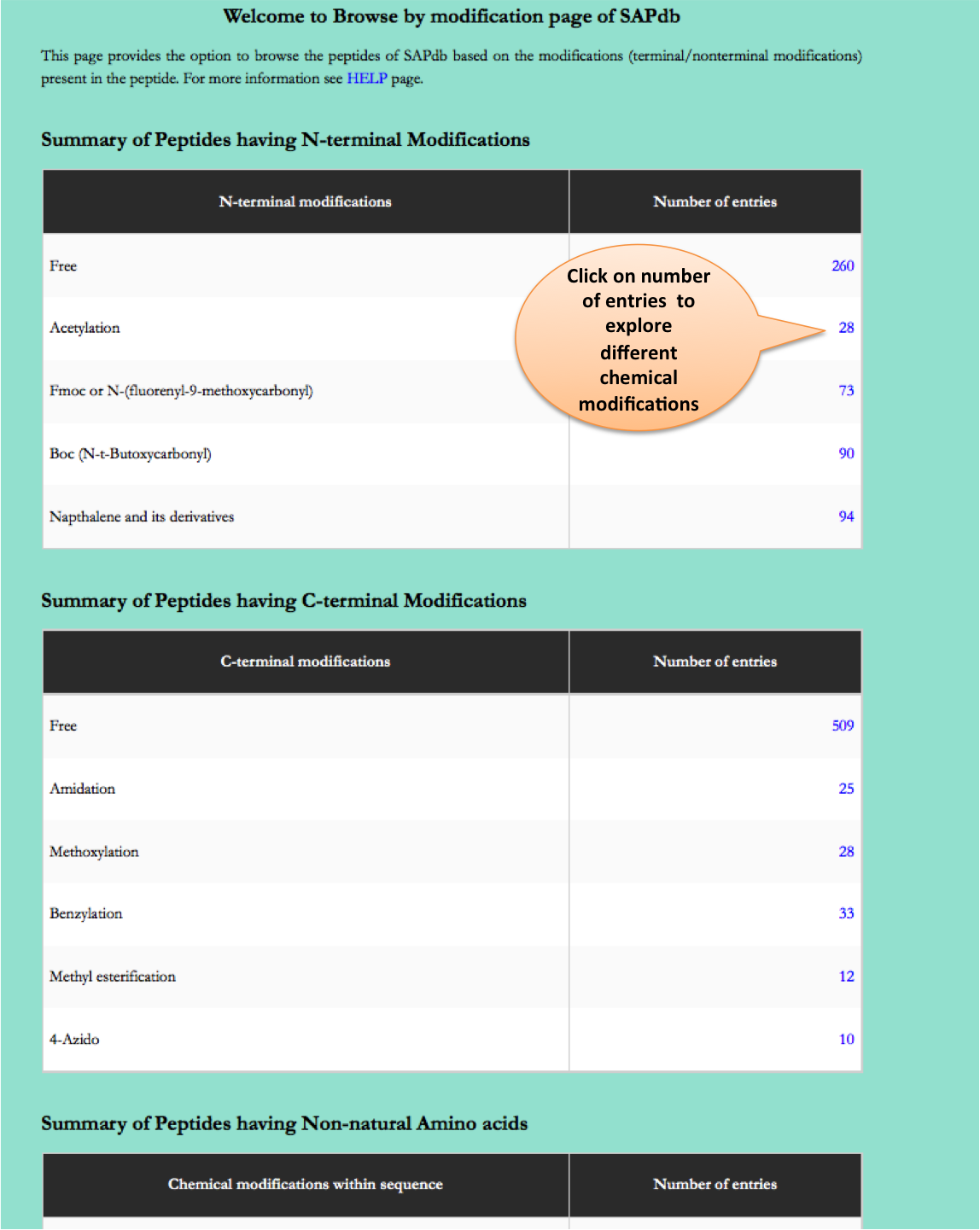
Chemical Modifications
This module displays the distribution of different modifications (N-terminal, C-terminal, Non terminal or non-natural amino acids) present in peptides present of the SAPdb.
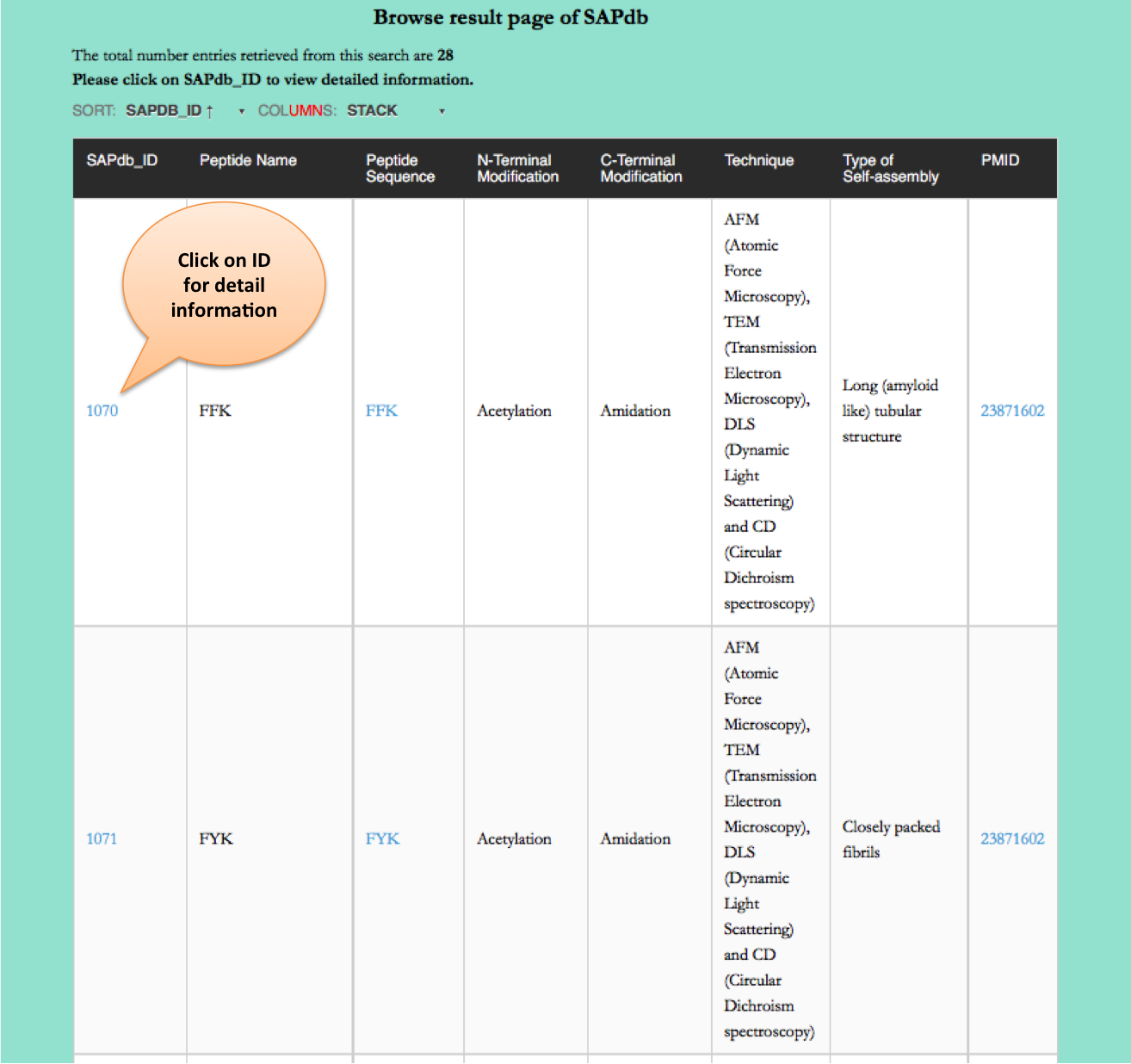
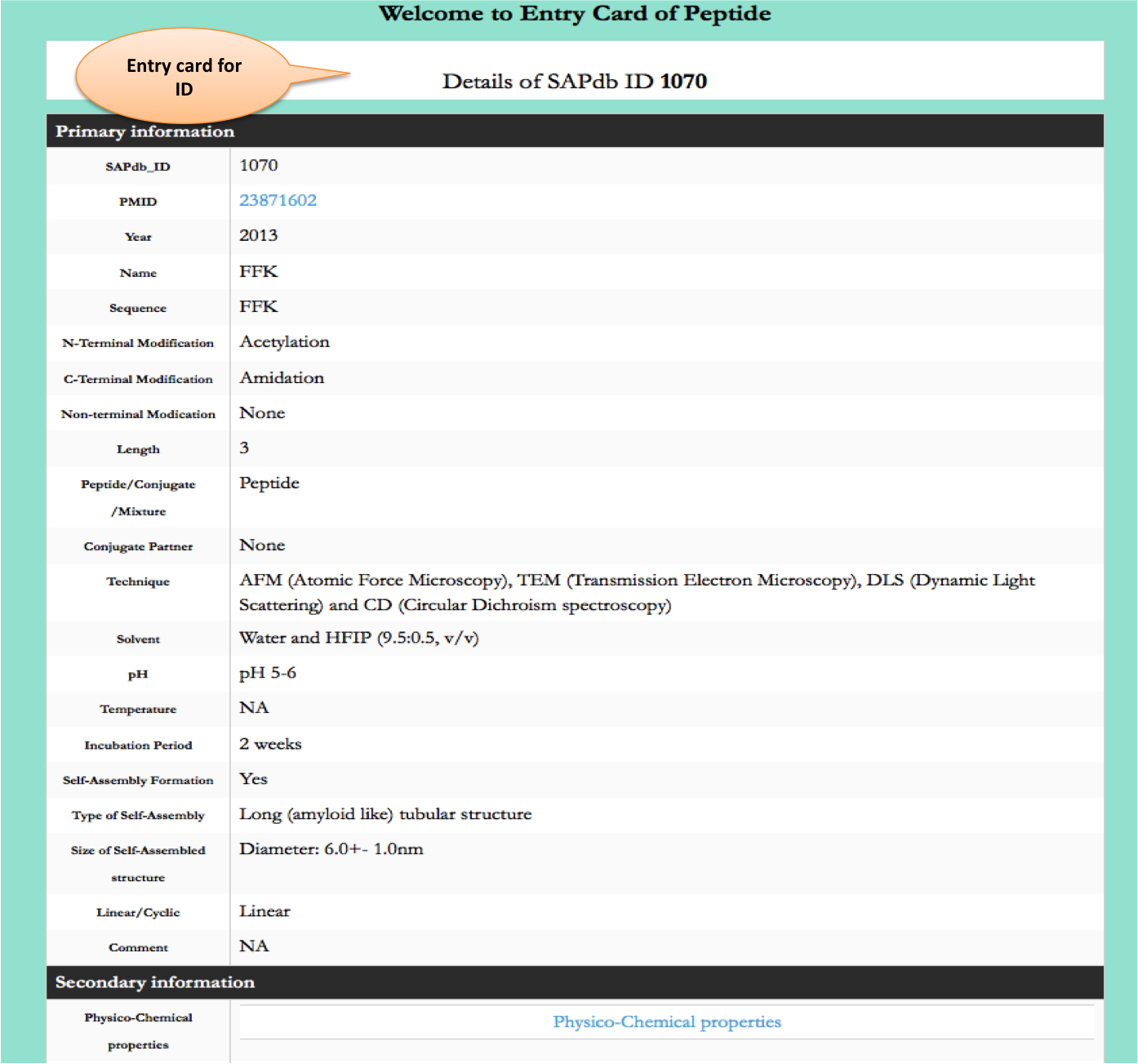
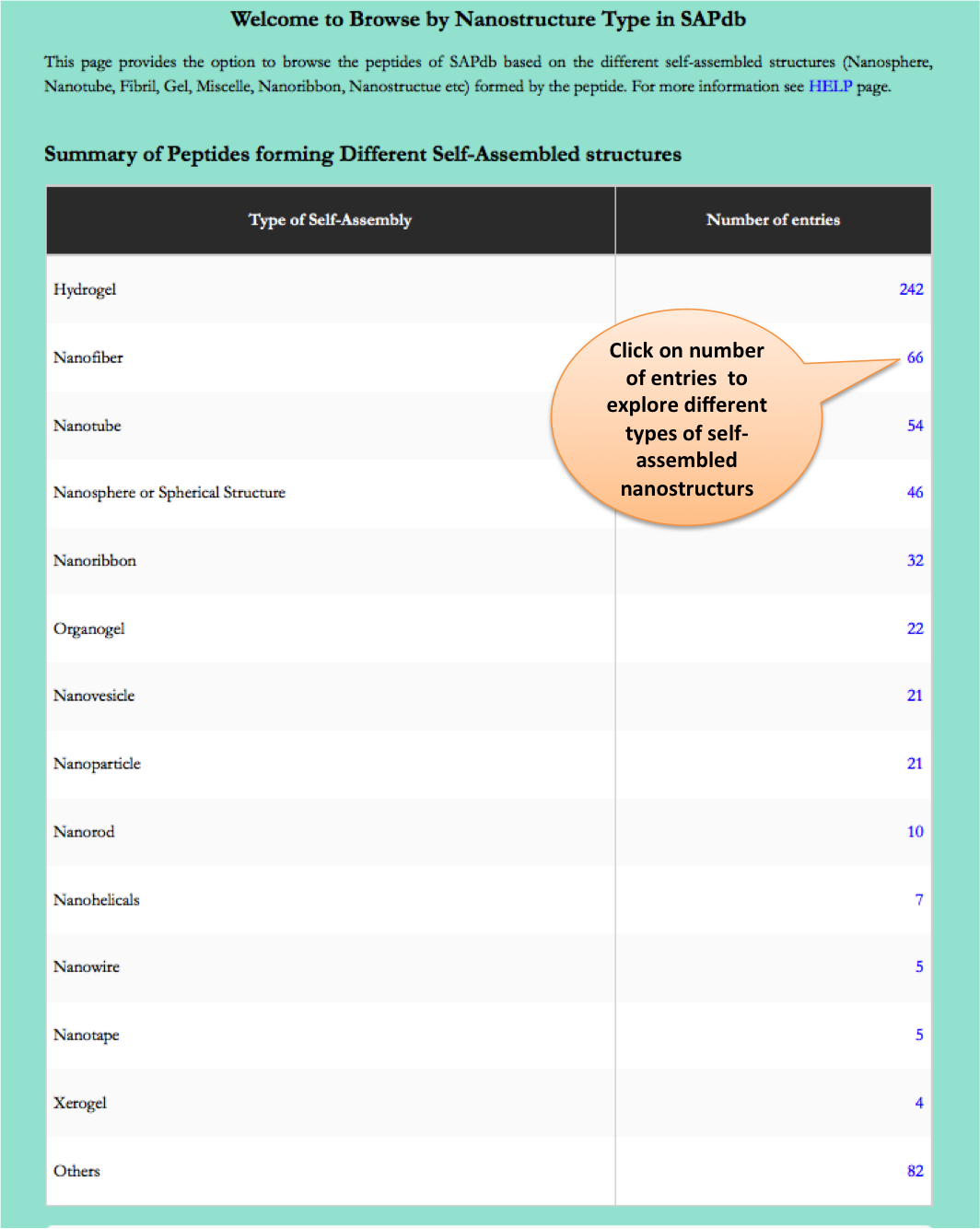
Type of Nano-structures
This page provides the distribution of of peptides on the basis of different nanostructres formed by peptides in SAPdb.
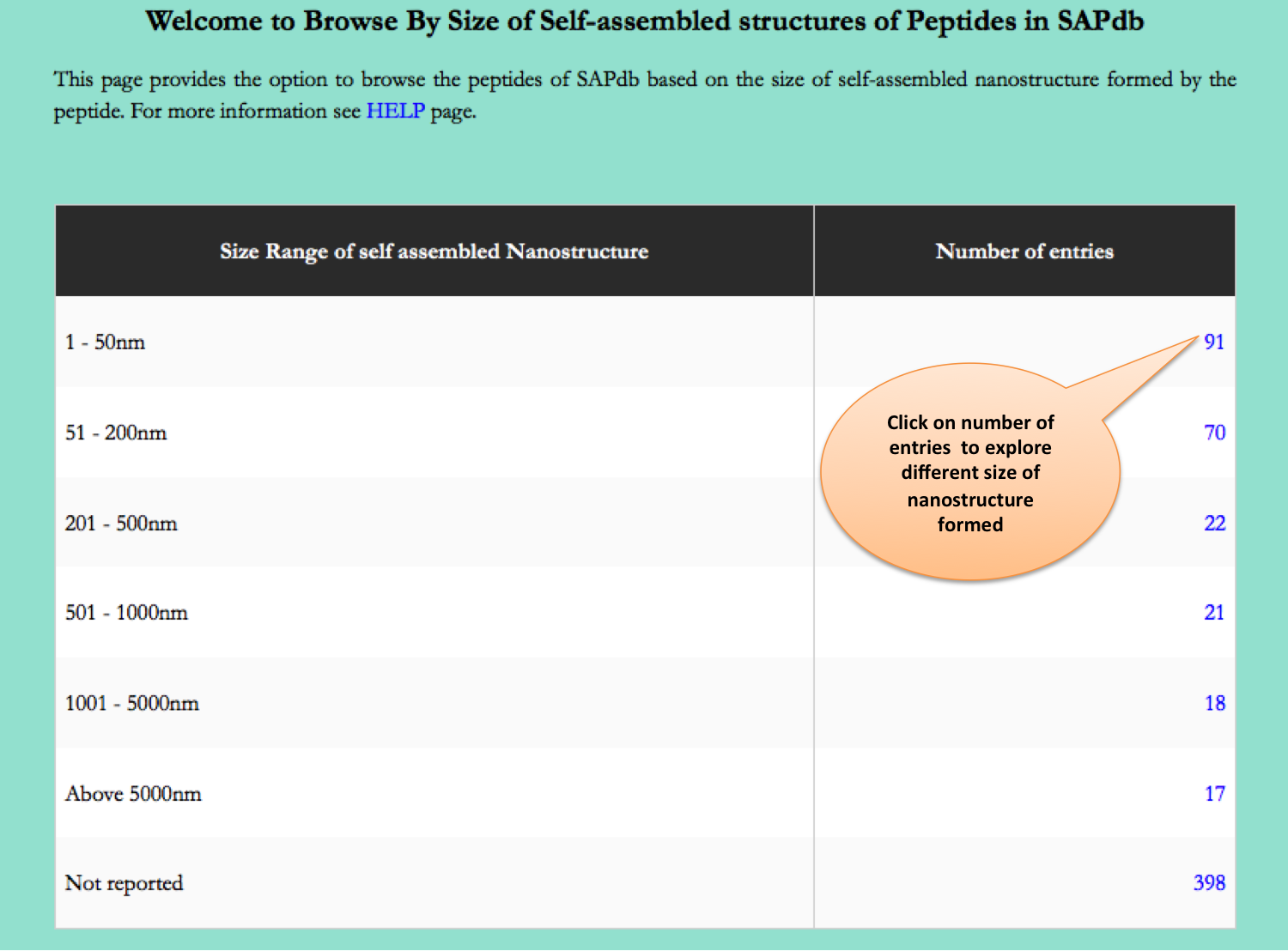
Size of Nanostructure
This page provides the distribution of of peptides on the basis of size of nanostructres formed by peptides in SAPdb.
Technique
This page provides the distribution of of peptides on the basis of itechniques used to analyze the nanostructres formed by peptides in SAPdb.

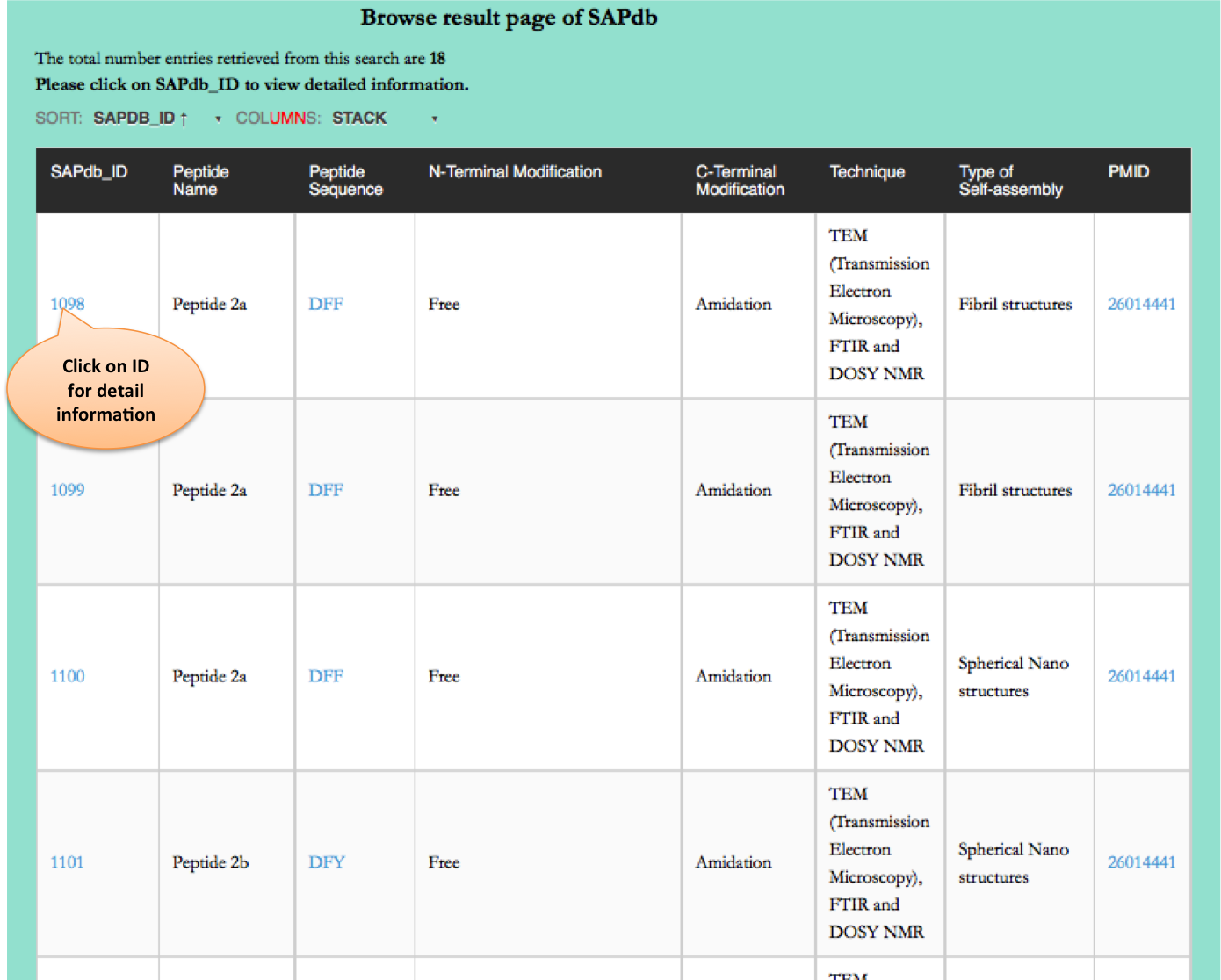

Dipeptide
This page provides the distribution of of peptides on the basis of dipeptide forming self-assembled nanostructures in SAPdb.

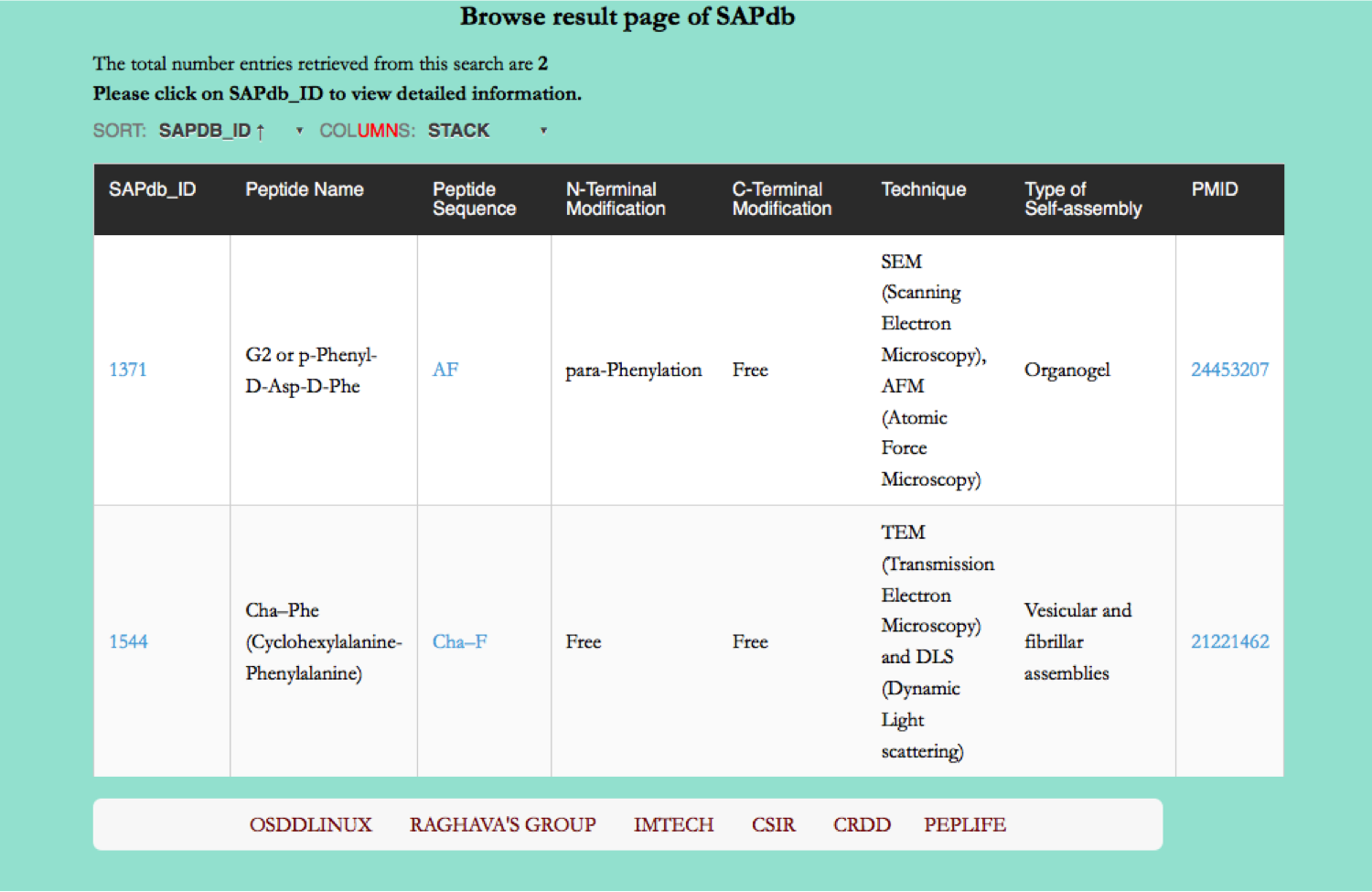
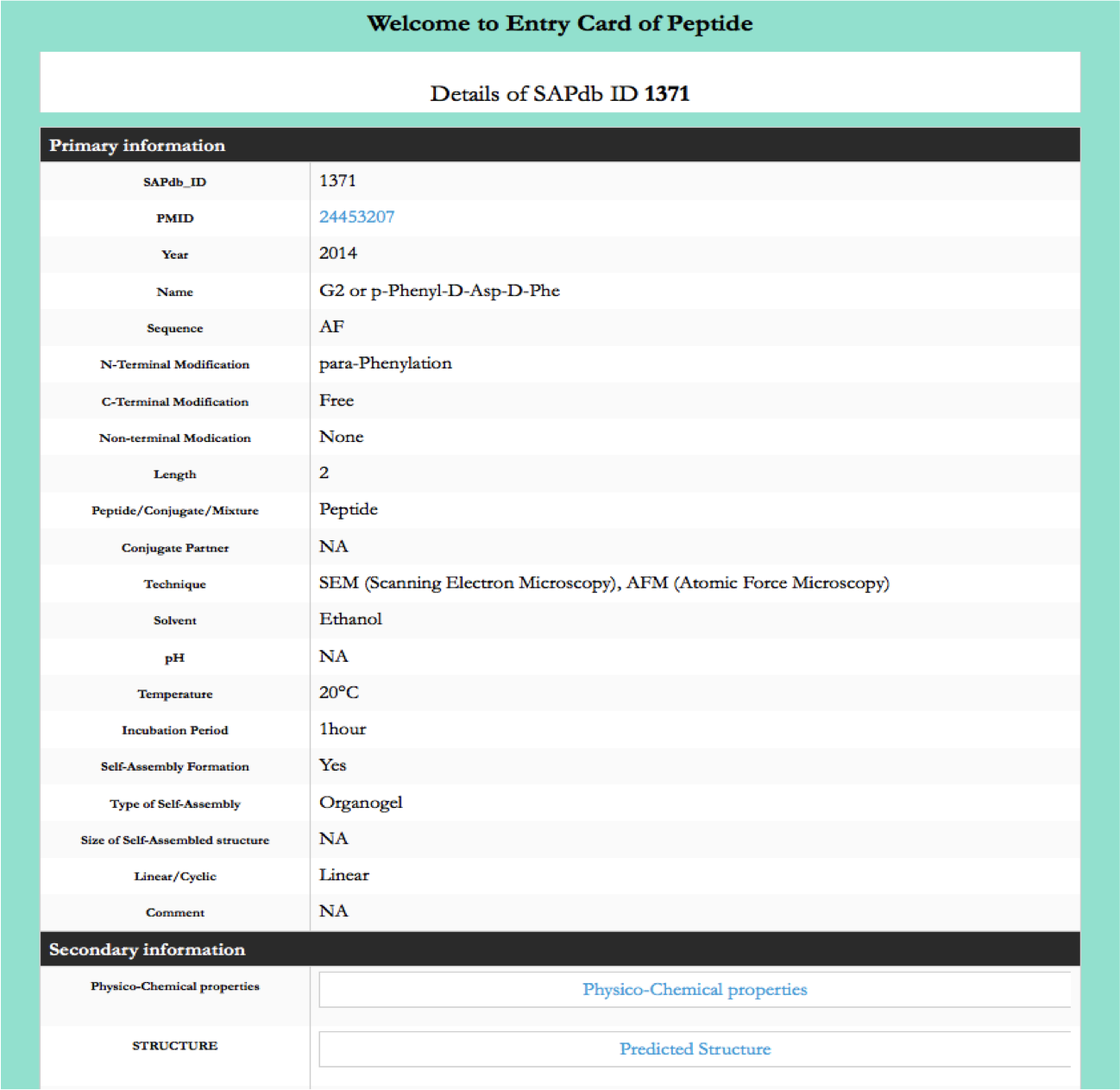
Tripeptides
This page provides the distribution of of peptides on the basis of tripeptide forming self-assembled nanostructures in SAPdb.
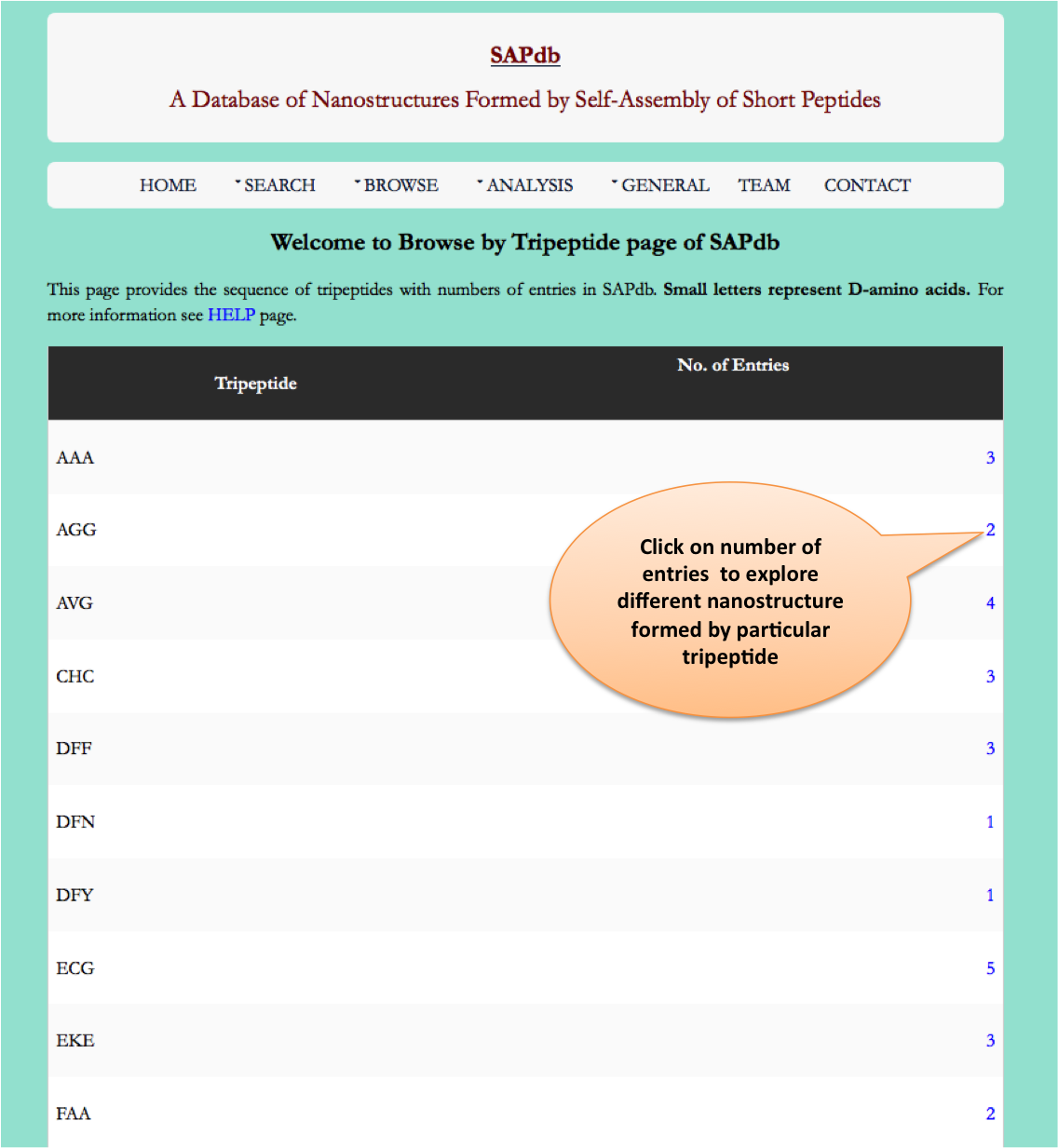
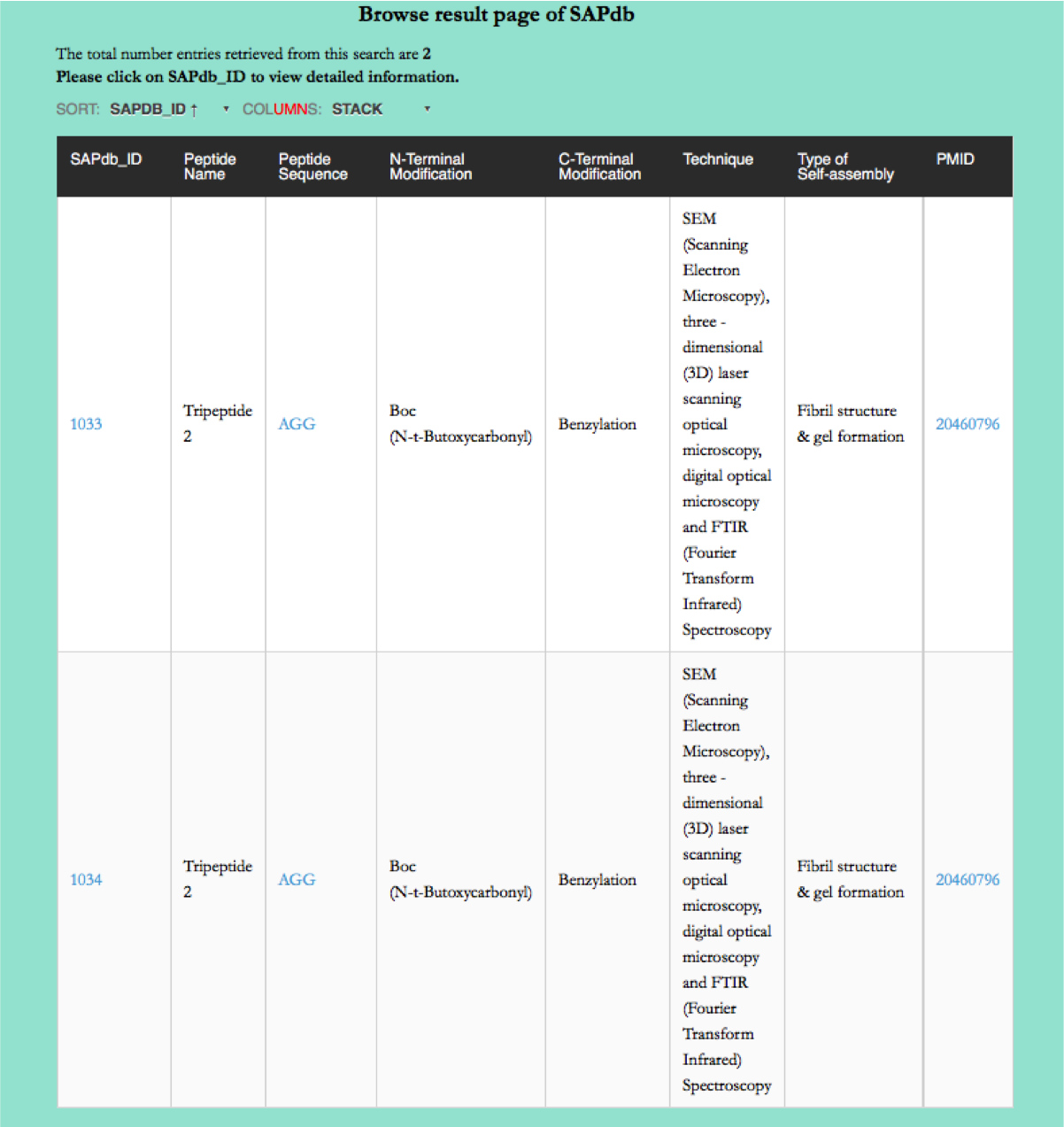
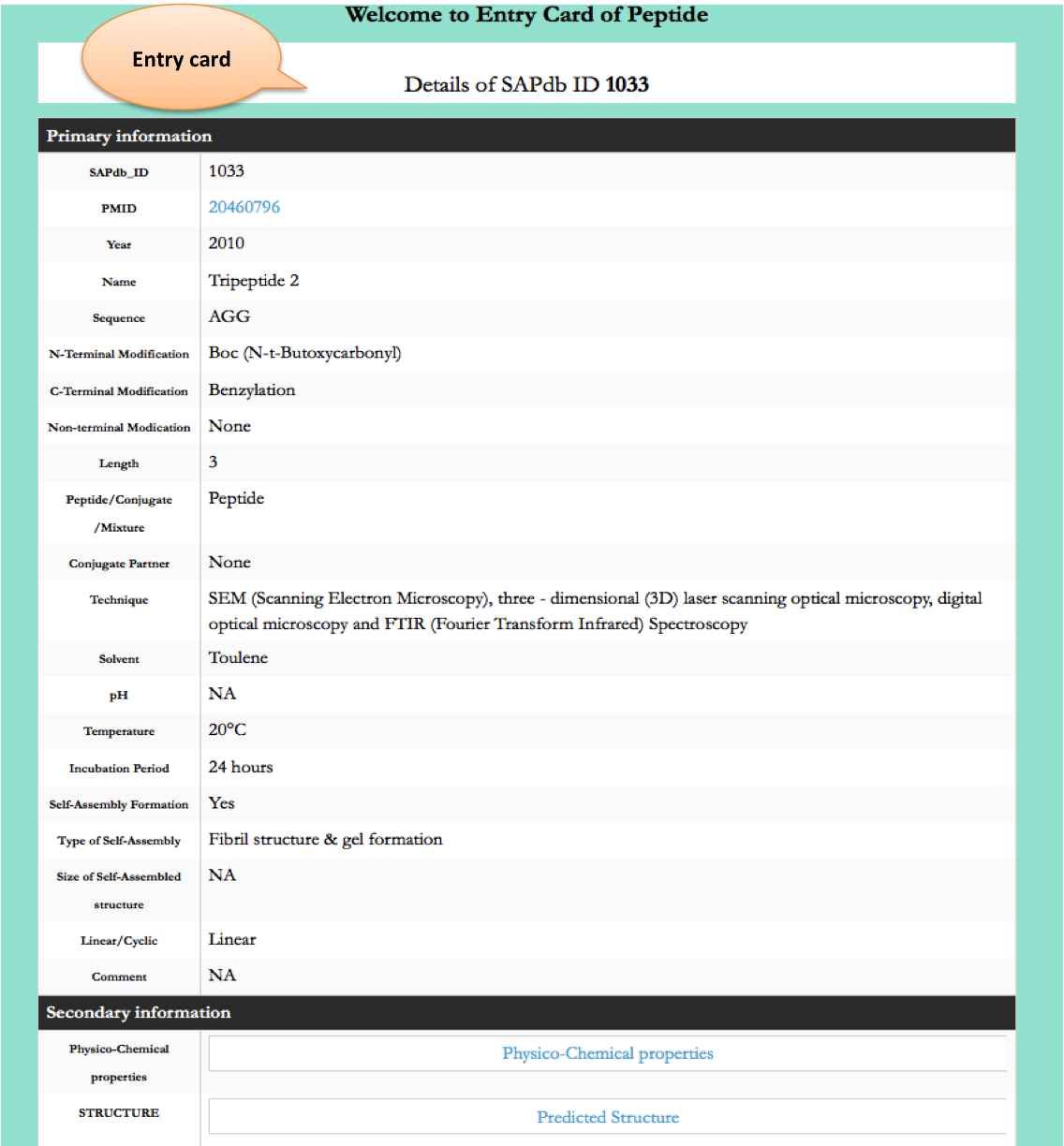
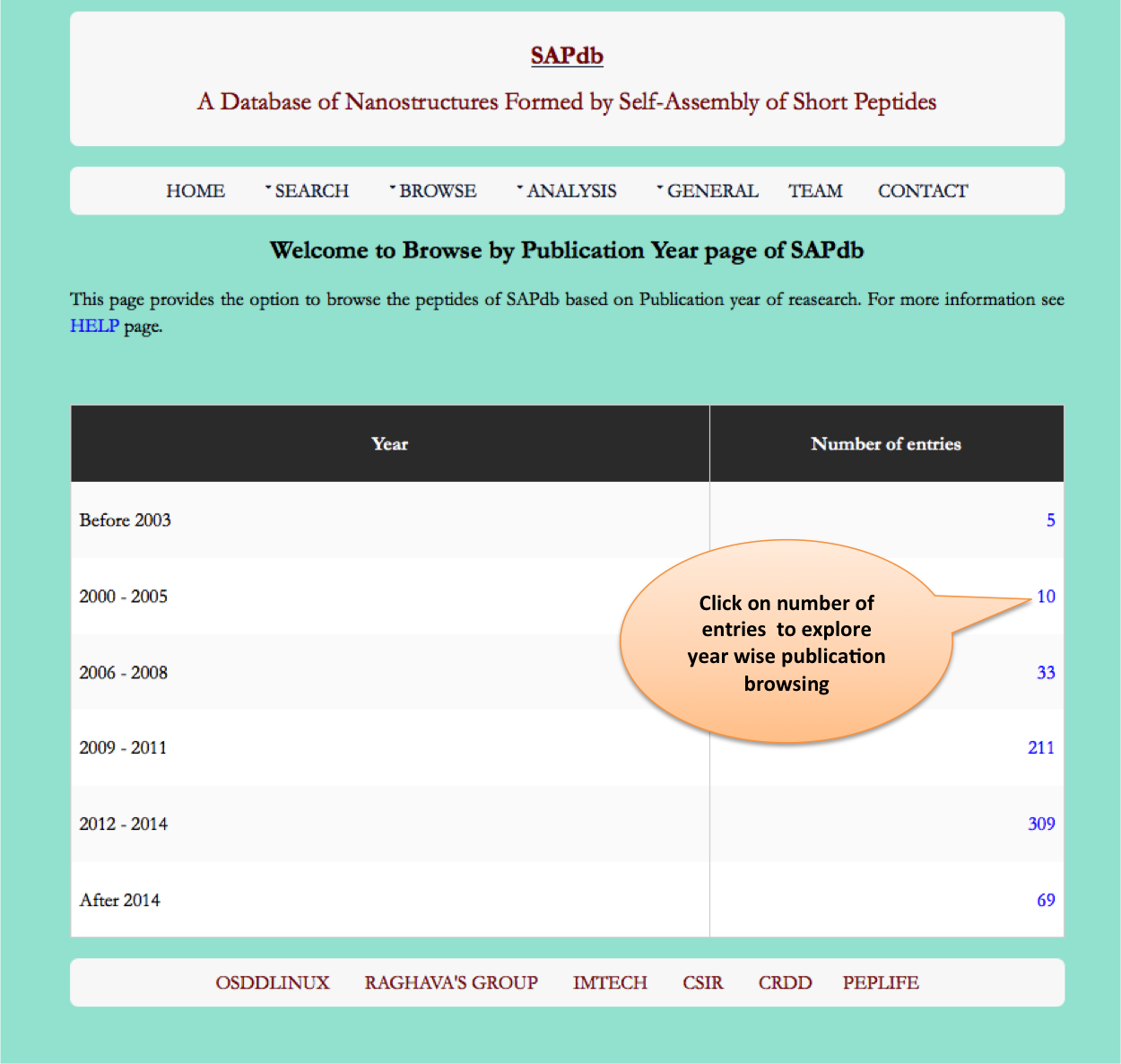
Year
This page provides the distribution of of peptides on the basis of year of publication from which entries has been made in SAPdb.
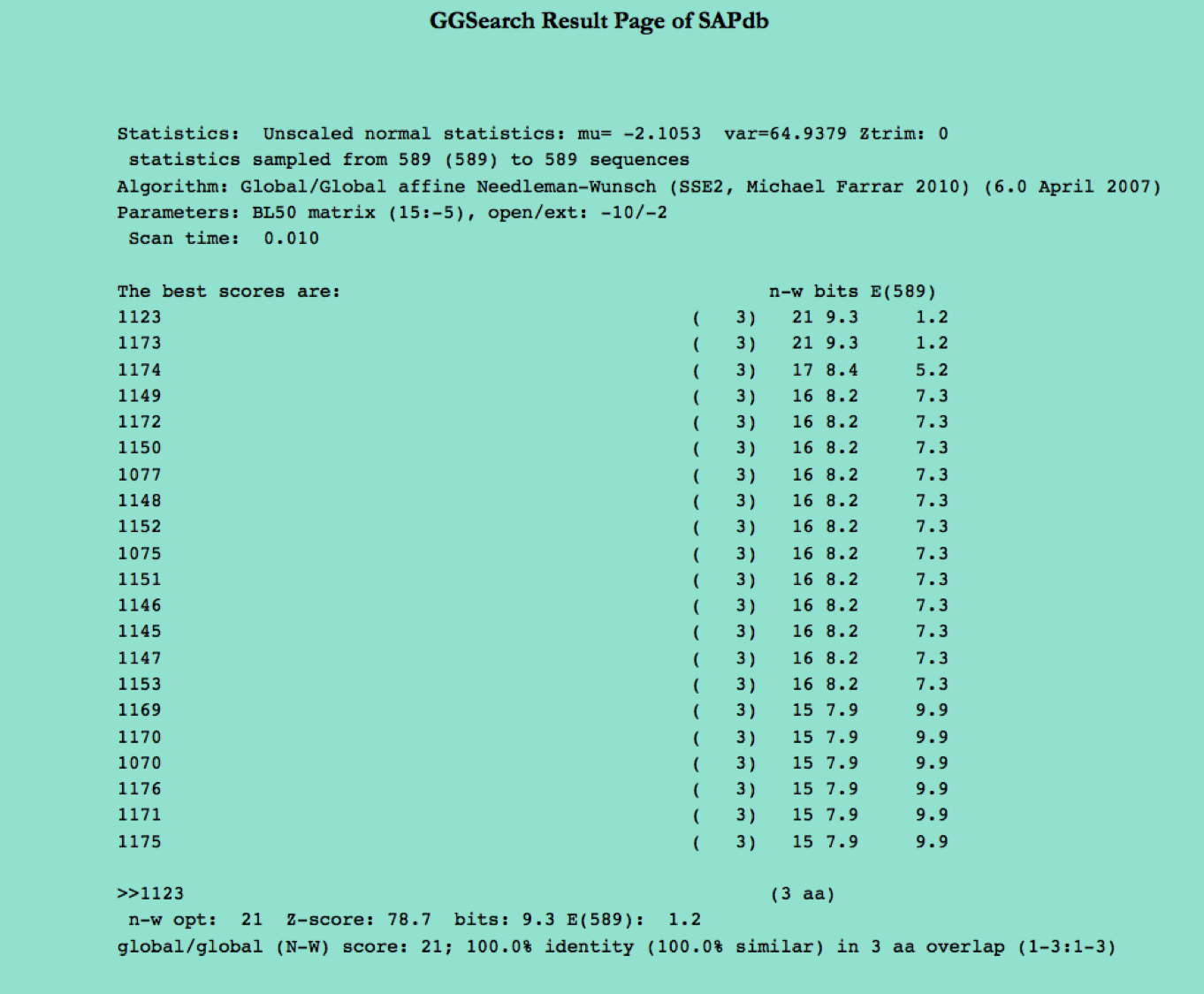
GGSearch
GGSearch page assists users in performing search against the peptides present in SAPdb. User can submit their peptide sequence in fasta format to performing similarity, server will returns the GGSearch output containing list of peptides similar to the query peptide.

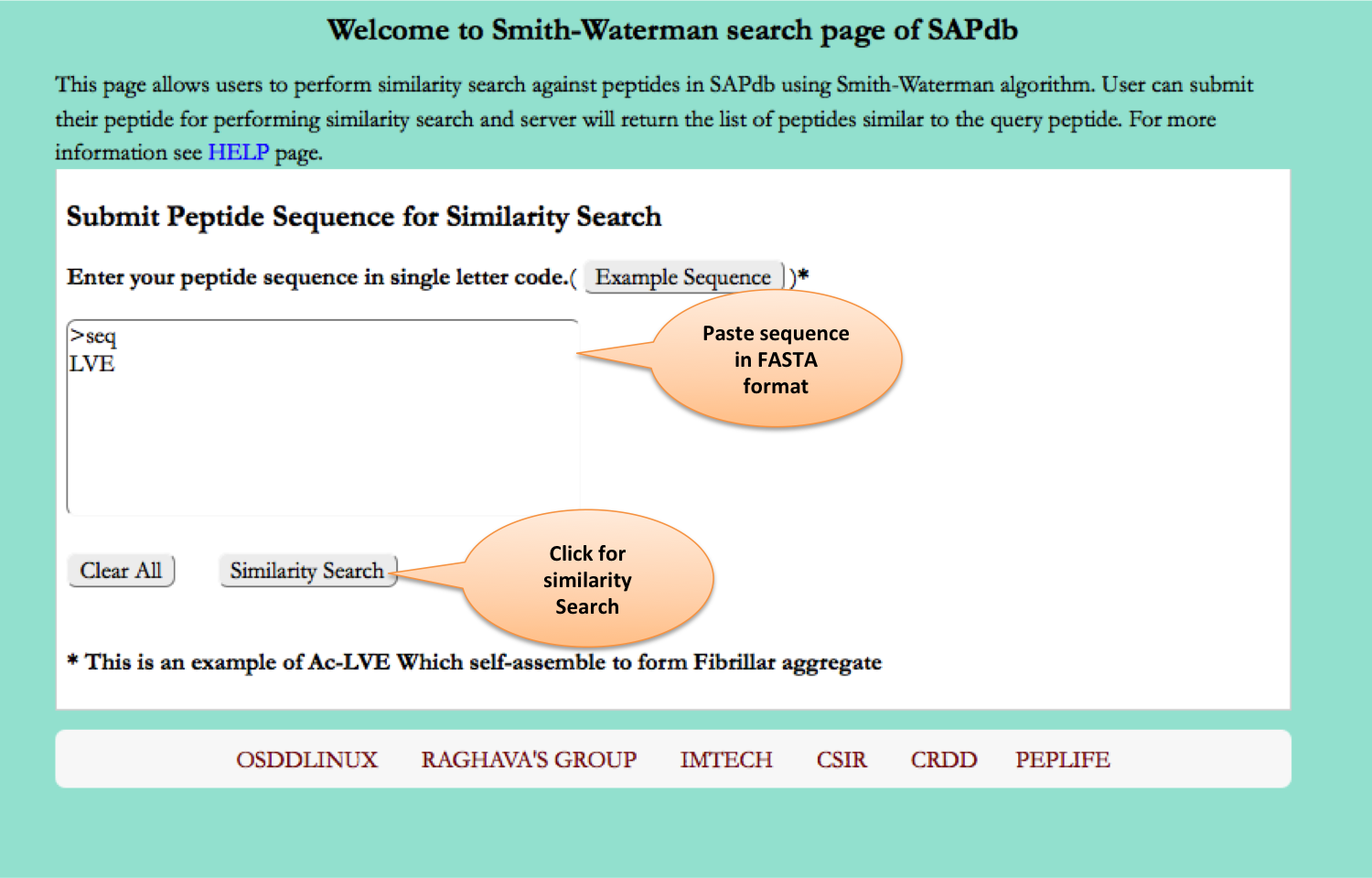
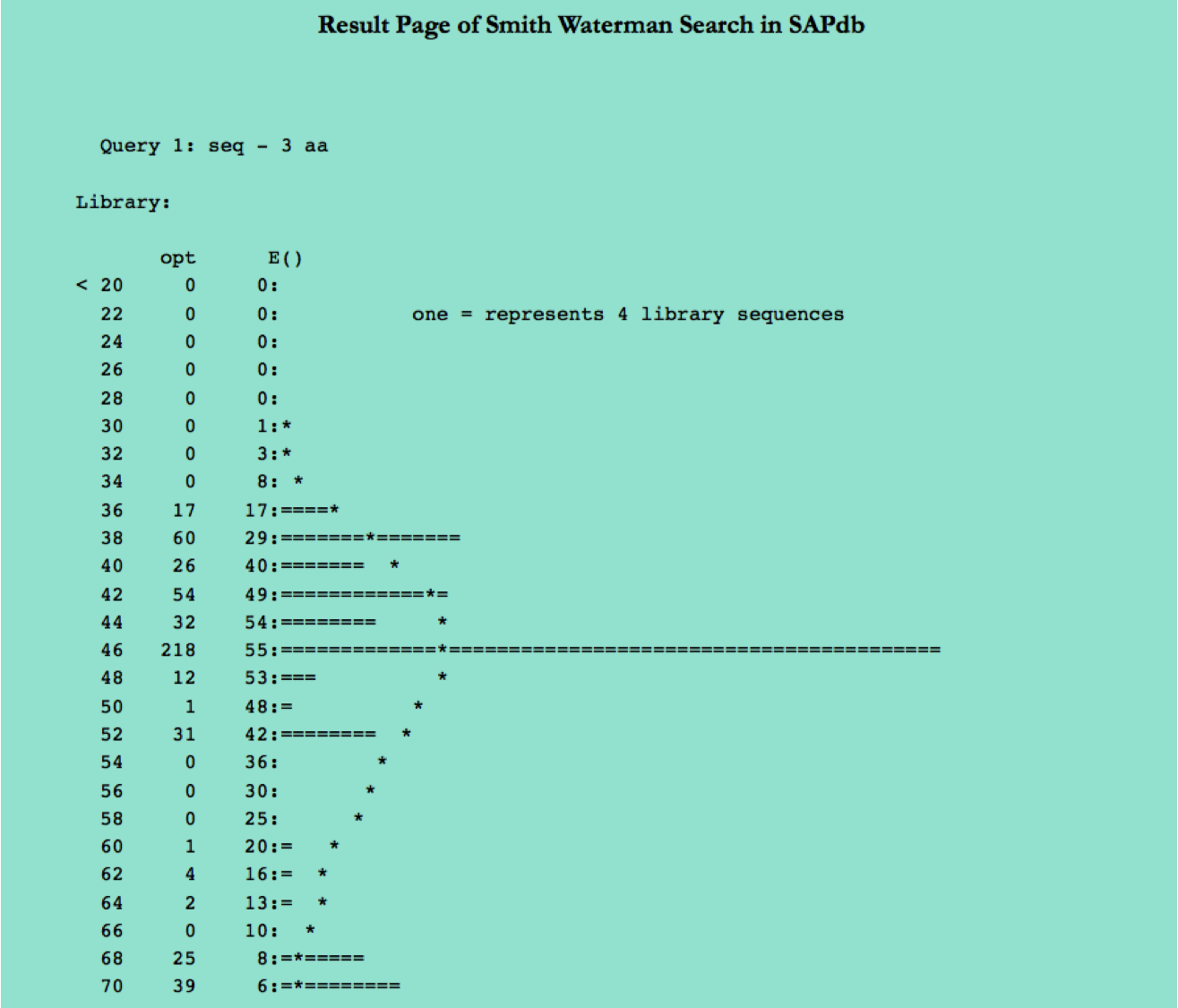
Smith-Waterman
This page allows users to perform similarity search against peptides present in SAPdb using Smith-Waterman algorithm. User can submit their peptide; server will returns the list of peptides similar to the query peptide.
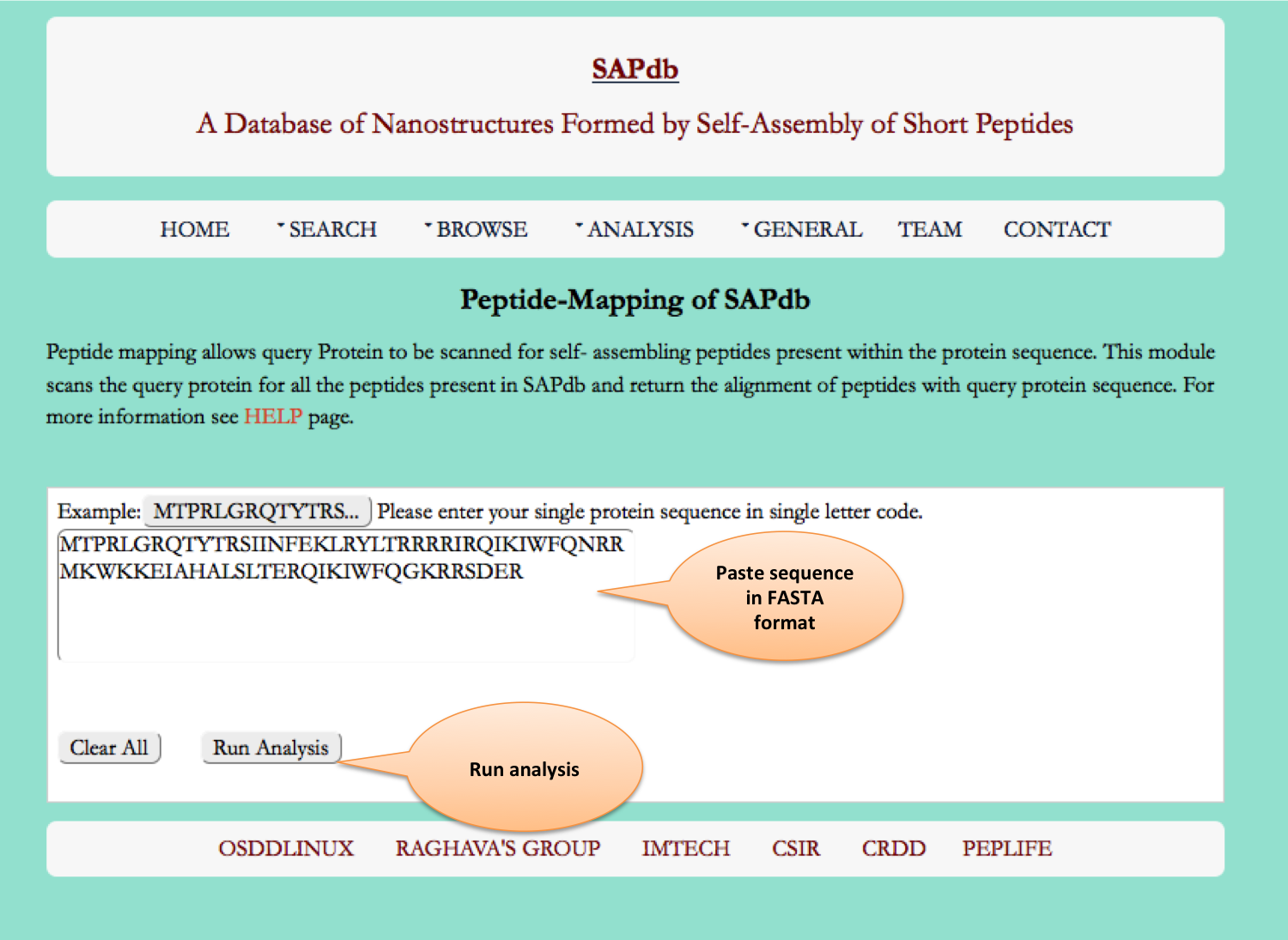
peptide mapping
Peptide Mapping module offers the user to run a sub-search and super-search. In sub-search a given peptide is mapped against all peptides of SAPdb Database. While super-search returns similar peptides of our database against protein sequence given as query.

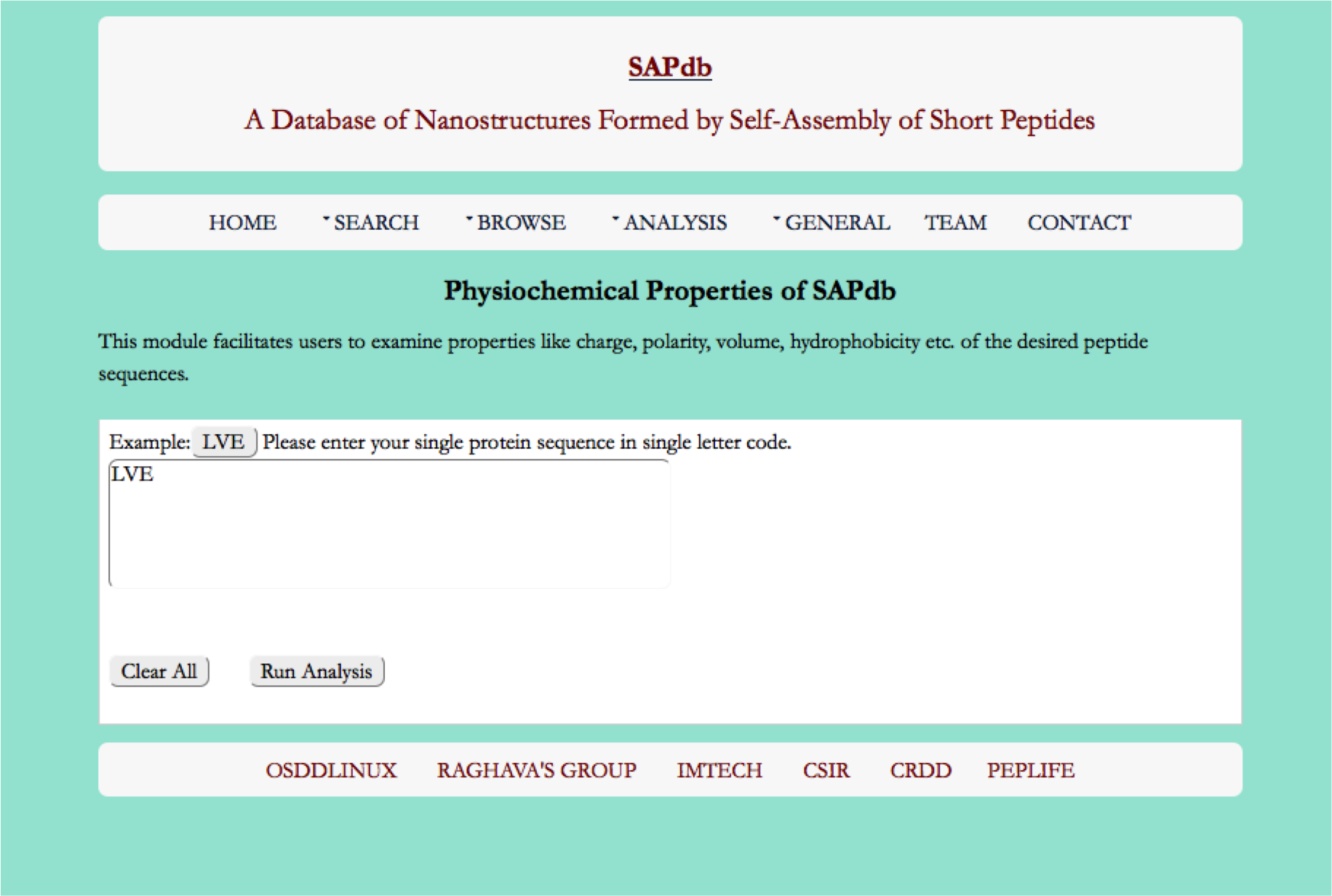
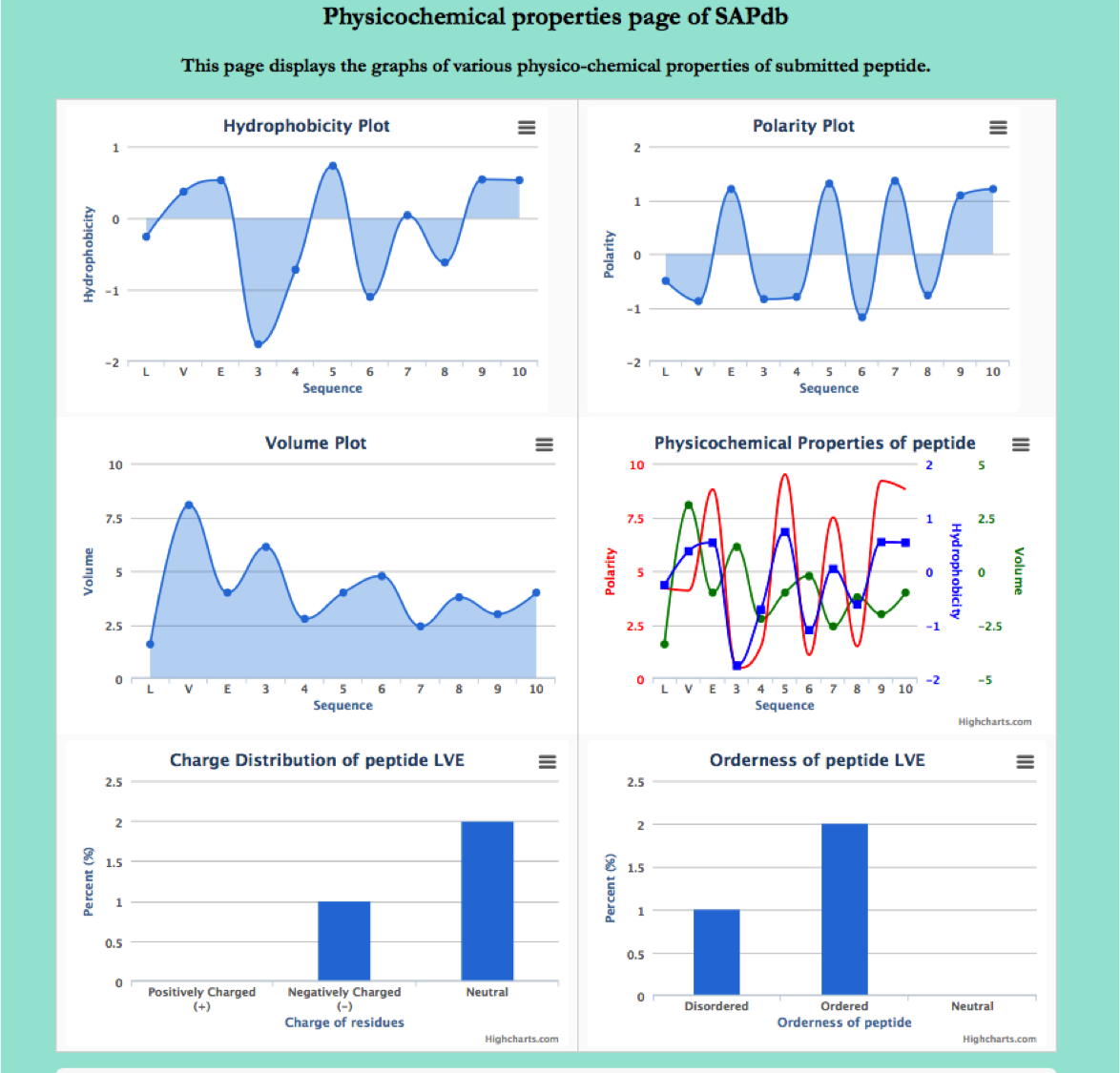
Physio-Chemical Properties
This Modules provides the physio-chemical properties (hydrophobicity, polarity, charge and orderness etc) in term of highchart for the peptides in SAPdb
Frequently Asked Questions (FAQs) |
|
Q1. What is SAPdb? Ans. SAPdb is an comrehensive resource that maintains dipeptides and tripeptides which form various self-assembled nanostructures. Q2. Why SAPdb? Ans. Dipeptides and tripeptides are the smallest peptides that can assemble into highly organized or ordered nanostructures. These are considered as ideal candidates to uncover the mechanism of self-assembly. Nanostructures formed by these small peptides have diverse applications in various arena such as building blocks for scaffolds in tissue engineering, drug delivery vehicles and biosensors etc. SAPdb is a resource, where we collect and organize comprehensive information (otherwise sccatered)regarding these Self-assembling small peptides that form defined nanostructures include sequence, modifications of peptides, conditions under which they form nanostructures, type of nanostructure and size of nanostructure. Thus SAPdb helps the researchers to attain a better understanding of the properties of peptides governing self-assembly of peptides. Q3. How to search into SAPdb? Ans. User can search SAPdb using various modules such as Search, Browse etc. In addition for sequence similarity search, user can explore various analysis tools that are integrated in SAPdb. |
