|
 |
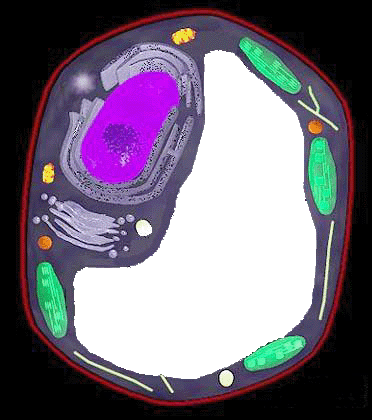
The recently sequenced rice genome has opened new challenges ahead for the plant research community in terms of identifying the function and regulation of each encoded protein. Gaining an understanding of the biological functions of novel genes is a more ambitious goal than obtaining just their sequences. To narrow this huge gap between the enormous amount of raw sequence data of the rice genome and the experimental characterization of the corresponding proteins, Professors therefore have to find computational ways to efficiently analyze these data. Subcellular location of a protein is one of the key functional characters as proteins must be localized correctly at the subcellular level to have normal biological function. We have therefore, developed RSLpred which is a SVM based prediction method for 4 major target proteins (Chloroplast, Cytoplasm, Mitochondria and Nucleus).
Name of Protein: This is an optional field. The name of protein may have letters and numbers with the "-" or "_". All other characters are non-permissible. The field is assigned a default name "Protein". The sequence name is just used for only your information. User may or may not enter the name of the sequence.
Paste a Protein Sequence: This server allows the submission of sequence in any of the standard formats. The user can paste plain sequence in the provided box. Amino acid sequences must be entered in the one-letter code. All the non-standard characters will be ignored from the sequence.
Upload Sequence File: The server also has the facility for uploading the local sequence files by using the "BROWSE" option.
Select Sequence Format: RSLpred can accept both the formatted or unformatted protein sequences. It uses ReadSeq routine to parse the input. The user should check the format of the input sequence before submitting the sequence for prediction. The results of the prediction will be false if the format choosen is wrong.
Choose a Prediction Approach: In case of default prediction, RSLpred uses the PSSM-based module. However, in the present investigation, we have developed 13 different SVM modules for the prediction of subcellular localization of rice proteins (see text for details). The best performing modules have been implemented on the World Wide Web as a dynamic web server where the Users have the option to choose either of the prediction approach available. The brief account of these approaches is given below: