ParaPep -A Database of Anti-parasitic peptides
ParaPep -A Database of Anti-parasitic peptides
HELP PAGE OF ParaPep | |
Parasite types and diseases they are causing |
| Parasite Type | Disease | Description |
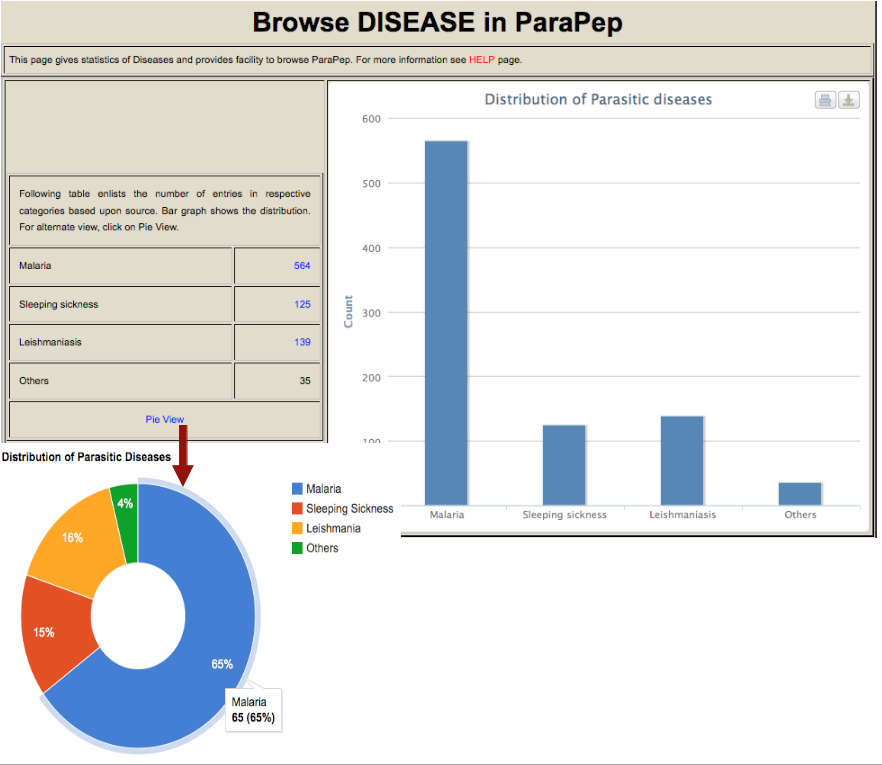
| Plasmodium | Malaria | Disease transmitted by female Anopheles mosquito. P.falciparum has biphasic life cycle- Mosquito and Human (Exoerythrocytic and Erythrocytic). Five species infect humans, P.falciparum most deadly. Artemisinin Combination Therapy(ACT) used for treatment. |
| Leishmania | Leishmaniasis | Female sandly transmit parasite. Obligate intra-macrophage single-celled protozoan. 20 species of Leishmania infect humans. Possible medications are liposomal amphotericin B, paromomycin, fluconazole. |
| Trypanosoma | Trypanosomiasis | Parasite transmitted to host by Tsetse fly (Sleeping Sickness).T.brucei, T.cruzi mainly infect humans. Diminazene, homidium, isometadium, suramin, and melarsomine used to treat infections. |
| Toxoplasma | Toxoplasmosi | Eating undercooked, contaminated food and water mainly causes infection. Parasites mainly form cysts in skeletal muscle, myocardium, brain. Medications include-Pyrimethamine, Clindamycin, Spiramycin, Sulfadiazine |
| Entamoeba | Amoebiasi | Transmitted by the fecal-oral route. Ingestion of cyst form of parasite causes infection. Symptoms range from mild diarrhea to severe dysentery. Metronidazole used for treatment. Boiled water used for drinking. |
| Babesia | Babesiosi | Usually transmitted by ticks. Parasite reproduce in red blood cells. Parasite require two host stick and mammal Oral atovaquone with oral azithromycin recommended for treatment. |
| Schistosoma | Schistosomiasi | Transmission of disease occurs through contamination of fresh water sources by excreta. It has trematode vertebrate-invertebrate lifecycle. People are infected by larval form of parasite. Drug praziquantel is used for treatment. |
| Caryospora | Pyrogranulomatous dermatiti | In dogs, pyogranulomatous skin lesions appear as papules, nodules. Treatment includes surgical excision of solitary nodules. Tetracycline and niacinamide used. |
| Neospora | Neosporosi | Neuromuscular disease of domestic dogs. It cycles between canine definitive hosts and herbivore intermediate hosts. Clindamycin (12.5-25 mg/kg PO or IM every 12 hours for 4 weeks) Trimethoprim sulfadiazine in combination with pyrimethamine is also used. |
| Besnoitia | Besnoitiosi | Transmission is through direct contact with animals with wounds. Life cycle complex with various intermediate hosts. Pedunculated lesions in the skin, nasal cavity and larynx of domestic animal are the major signs. |
| Eimeria | Hemorrhagic cecal coccidiosi | Acquired via fecal contamination of food and water. Monoxeous life cycle with the definitive (only) host as chickens. Morbidity is 10-40% and mortality up to 50%. Toltrazuril, Sulphonamides, Amprolium, Vitamins A and K in feed or water. |
| Cryptosporidium | Cryptosporidiosi | Spread through the fecal-oral route, often through contaminated water. Self-limiting diarrhea , stomach pains or cramps, low fever, nausea are the major symptoms. Nitazoxanide (FDA) is used for cure. paromomycin, atovaquone and azithromycin are also sometimes used. |
How to use ParaPep |
| This page provides help to the users and explains the working of each tool integrated in this database.Click on the links below to go to the help section of the respective tool. |
|