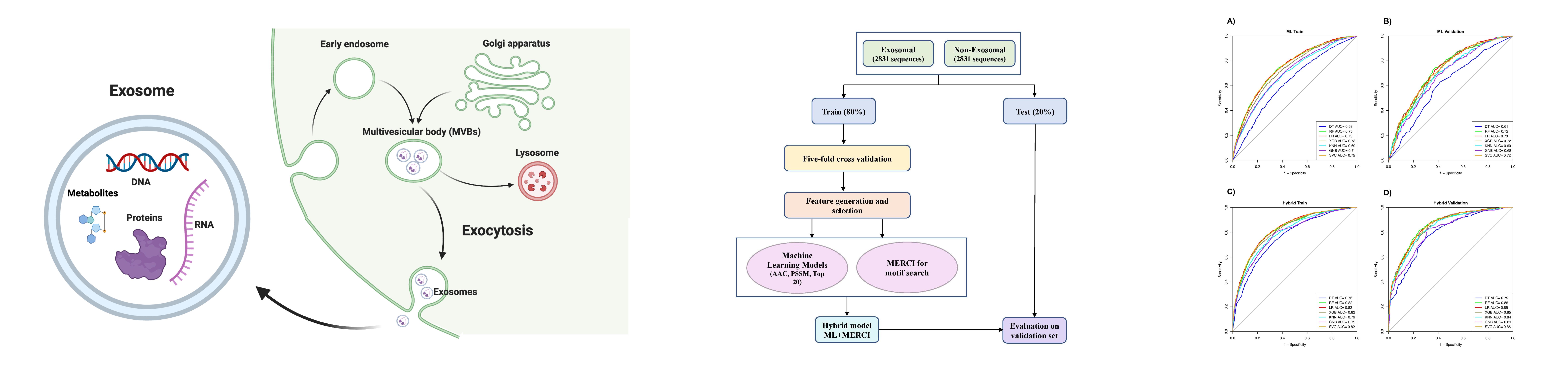
ExoProPred is a webserver to predict exosomal proteins based on hybrid model that combines machine learning model with motif-search approach. The models are trained on a dataset comprising of 2831 exosomal proteins and 2831 non-exosomal proteins. The performance of the models were evaluated using 5-fold cross-validation. The models were trained on top 70 best features comprising of composition-based and evolutionary information based features as well as on hybrid features(Top 70 features + Motif-search) by implementing random-tree classifiers from the scikit library of python. The datasets were extracted from Uniprot database (Uniprot). This method will be highly useful in the fields of development of non-invasive diagnostic methods, treatments, drug delivery system, designing personalized therapies, etc
Cite: Arora A, Patiyal S, Sharma N, Devi NL, Kaur D, Raghava GP (2023) A random forest model for predicting exosomal proteins using evolutionary information and motifs. Proteomics. e2300231. doi: 10.1002/pmic.202300231.