Welcome To The Algorithm Page of DefPred
| DefPred is a web-server with the capability of classifying defensins/non-defensins using AMPs and random protein sequences. Defensins are tiny cysteine-rich cationic proteins found in vertebrate and invertebrate mammals, plants, and fungi. They're host protective, and some of them have direct antimicrobial action, immune signalling activity, or both. Mammalian defensins are multifunctional, and through communicating with host cell receptor(s), they participate in both the host's innate and adaptive antimicrobial immunity, according to recent proof. In the past, numerous studies have highlighted the introduction of new potential Defensins and shown their functions in various diseases. The discovery of their role and structure is a crucial step in the development of therapies and vaccines. The prediction server for defensins has been developed in a very user-friendly manner. Here, on this page, user can get the details of all the algorithms and procedures exploited in the predict, design and protein scan module. |
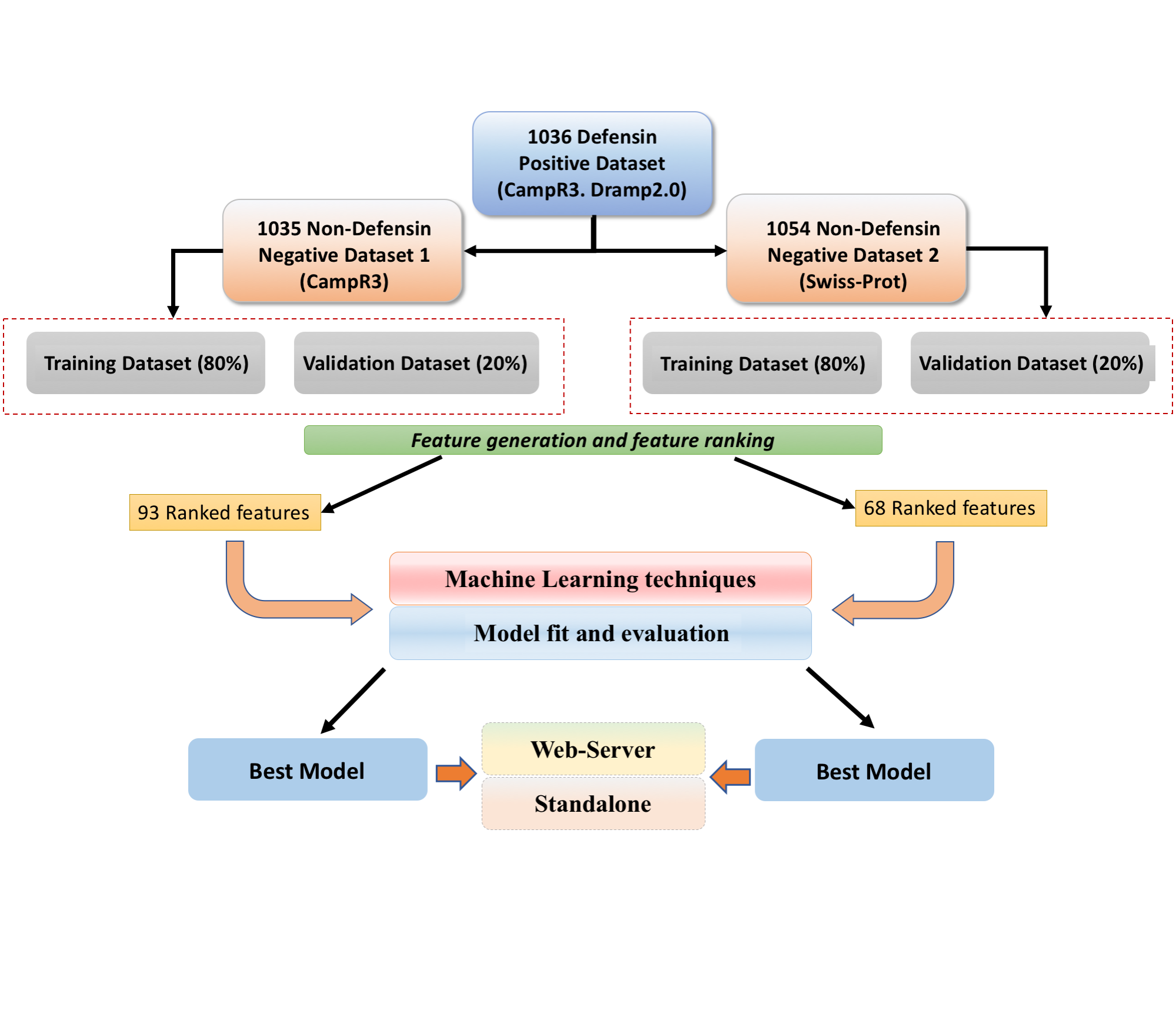
Datasets Used:In order to develop the prediction method, we have collected the datasets from two different sources, which are explained below.
|
The workflow shown in the above figure represents the model development for this server's three major modules..
Prediction moduleThe "Predict" module provides the facility to the user to classify defensins sequence(s) from antimicrobial pepties using Model 1 and from any random protein sequqnce using Model 2. User can provide multiple sequences as input to the server to predict the given sequence as defensins/non-defensins. In this study we used Support Vector Classifier to develop prediction modules.Design moduleThe "Design" module provides the facility to the user to create all possible analogs/mutants of the input sequence and classify into defensins sequence(s) from antimicrobial pepties using Model 1 and from any random protein sequqnce using Model 2. User can provide only one sequence at a time as input to the server to predict the given sequences as defensins/non-defensins. In this study we used Support Vector Classifier to develop prediction modules.Protein Scan moduleThe "Protein Scan" module provides the facility to the user to classify defensins sequence(s) from the non-defensins sequence(s) by generating the overlapping patterns of user specified length from the submitted sequence. User can provide only one sequence as input to the server to generate and predict the given sequence as defensins/non-defensins. |
