Welcome To The DefPred
Prediction of Defensins
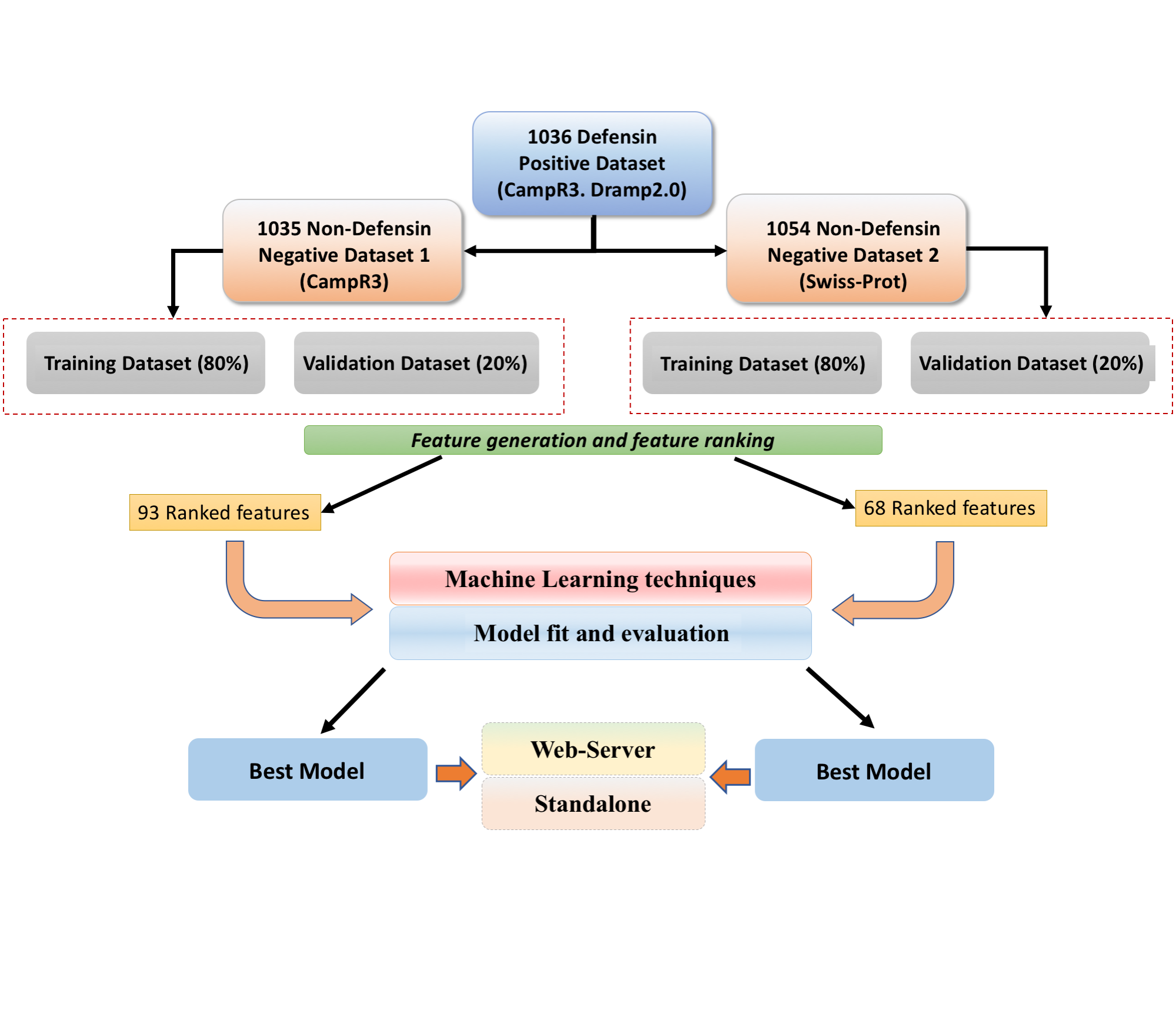
Defpred is a web server that predicts the presence of Defensins in given query protein sequqnces. Defensins are tiny cysteine-rich cationic proteins found in vertebrate and invertebrate mammals, plants, and fungi. They're host protective, and some of them have direct antimicrobial action, immune signalling activity, or both. Mammalian defensins are multifunctional, and through communicating with host cell receptor(s), they participate in both the host's innate and adaptive antimicrobial immunity, according to recent proof. In the past, numerous studies have highlighted the introduction of new potential Defensins and shown their functions in various diseases. The discovery of their role and structure is a crucial step in the development of therapies and vaccines. This method was created to classify and predict Defensins using the most extensive dataset available. Primary protein sequences ranging in length from 10 to 60 residues were used to develop the prediction modules using main and alternate datasets. We provided two models here Model-1 for prediction of defensin and antimicrobial peptides, model-2 for predicting defensin and non-defensin.