Top
DBpred Help Page
This is a help page of DBpred, developed for predicting DNA-Interacting residues in a protein. This page provides diffrent type of information on DBpred. In order to provide information in structured forms, we have divided information in following topics.
- Datasets: Datasets were generated from DNA-interacting protein structure in PDB (April 2019, release).
- Evaluation of models: Standard protocols were used for evaluating models develop in this study.
- Algorthm: Standard algorithm are used for devloping DNA interacting residues.
- Help on Web Pages: Help pages with screen shots
Goto Top
Goto Top
Goto Top
Goto Top
Goto Top
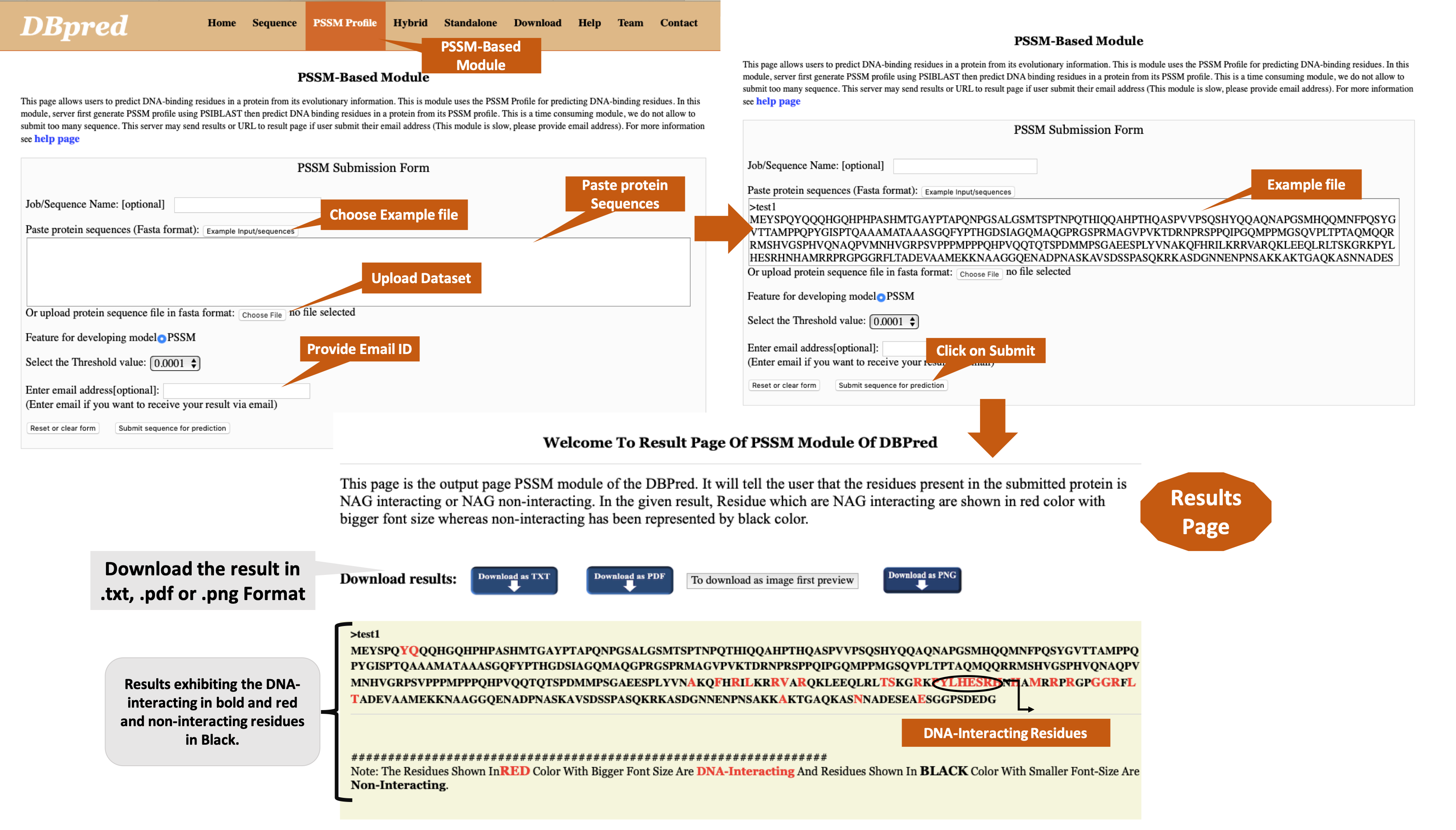
Help PagesDBpred server discriminate the DNA-Interacting residues and non-interacting residues from a given sequence. |
|
This page provides help on different modules of server. Following are major modules in this server:
|
Hybrid Profile: This server allow users to predict DNA binding residues in a protein from its primary structure information, by using the hybrid of three different features such as amino acid binary (AAB) profile, physico-chemical properties based binary (PCB) profile, and Position-Specific Scoring Matrix (PSSM) profile. This module allow users to submit multiple protein sequences at a time in FASTA format.