Welcome to Help Page
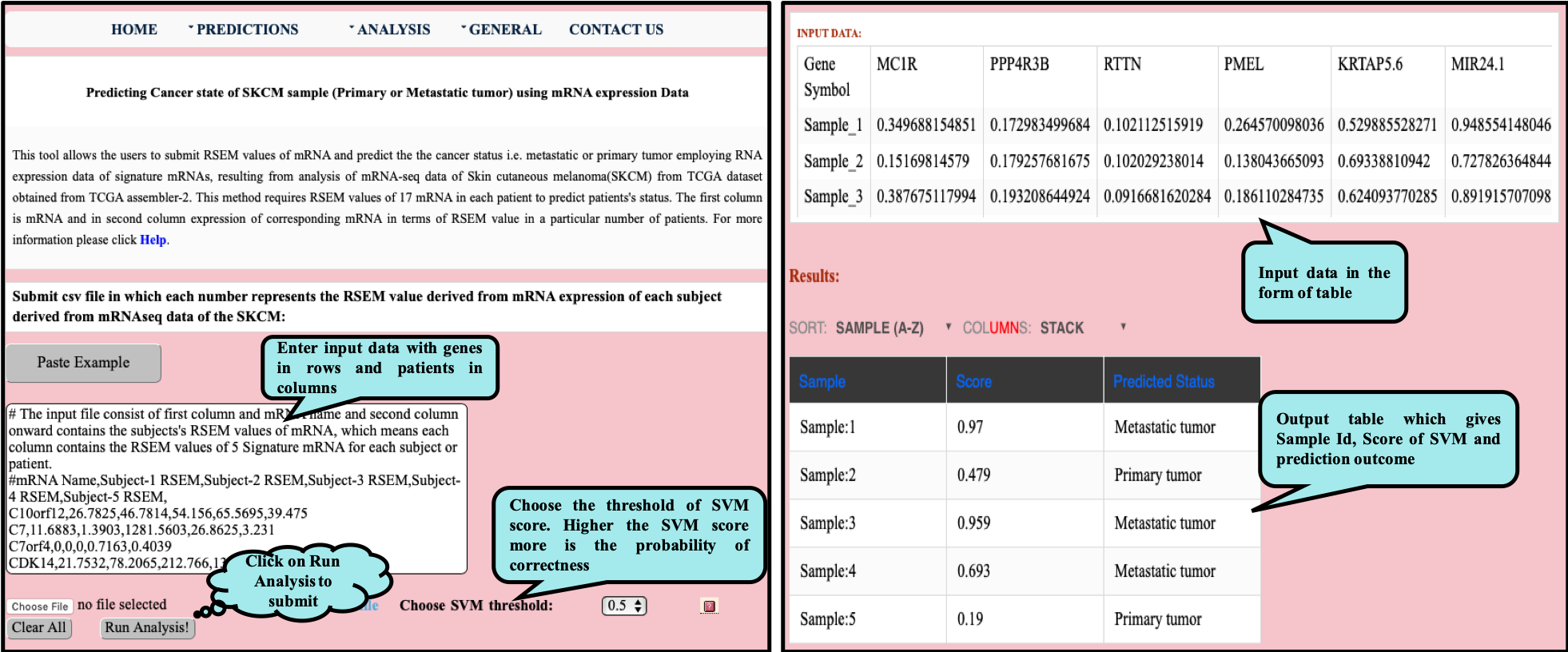
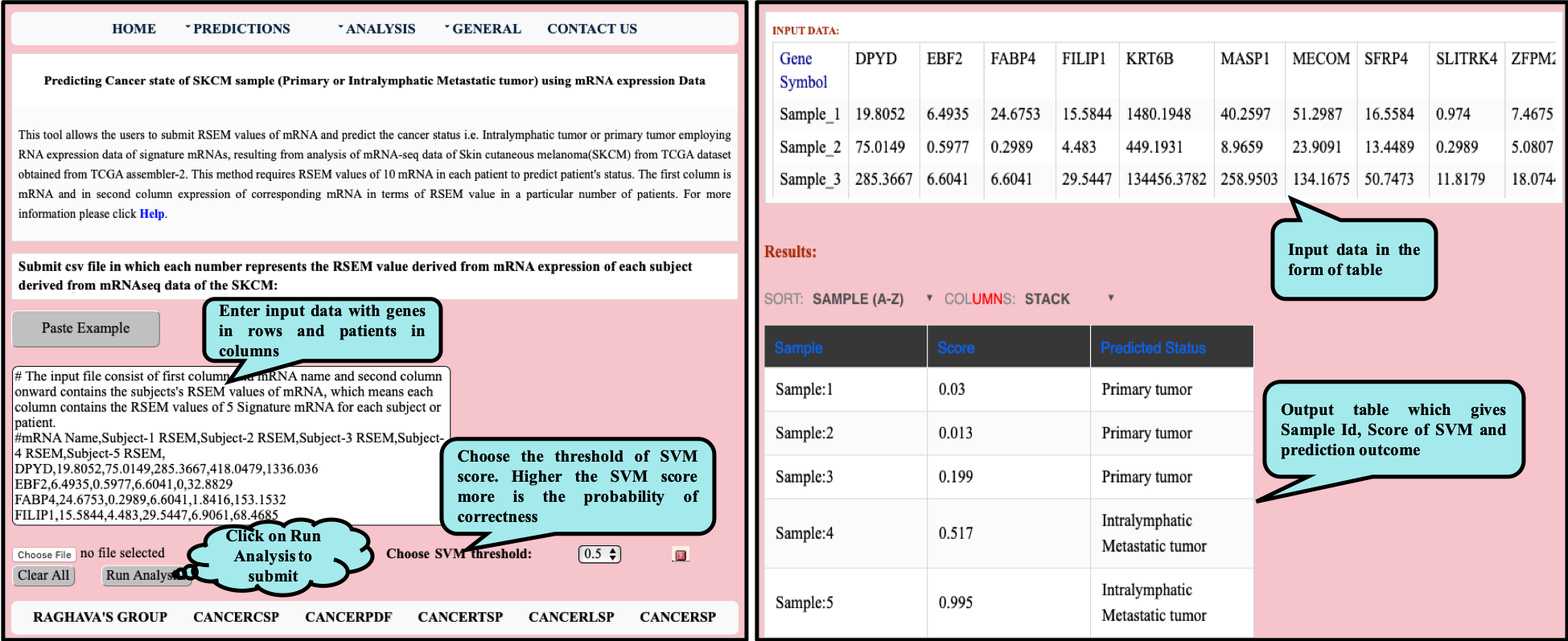
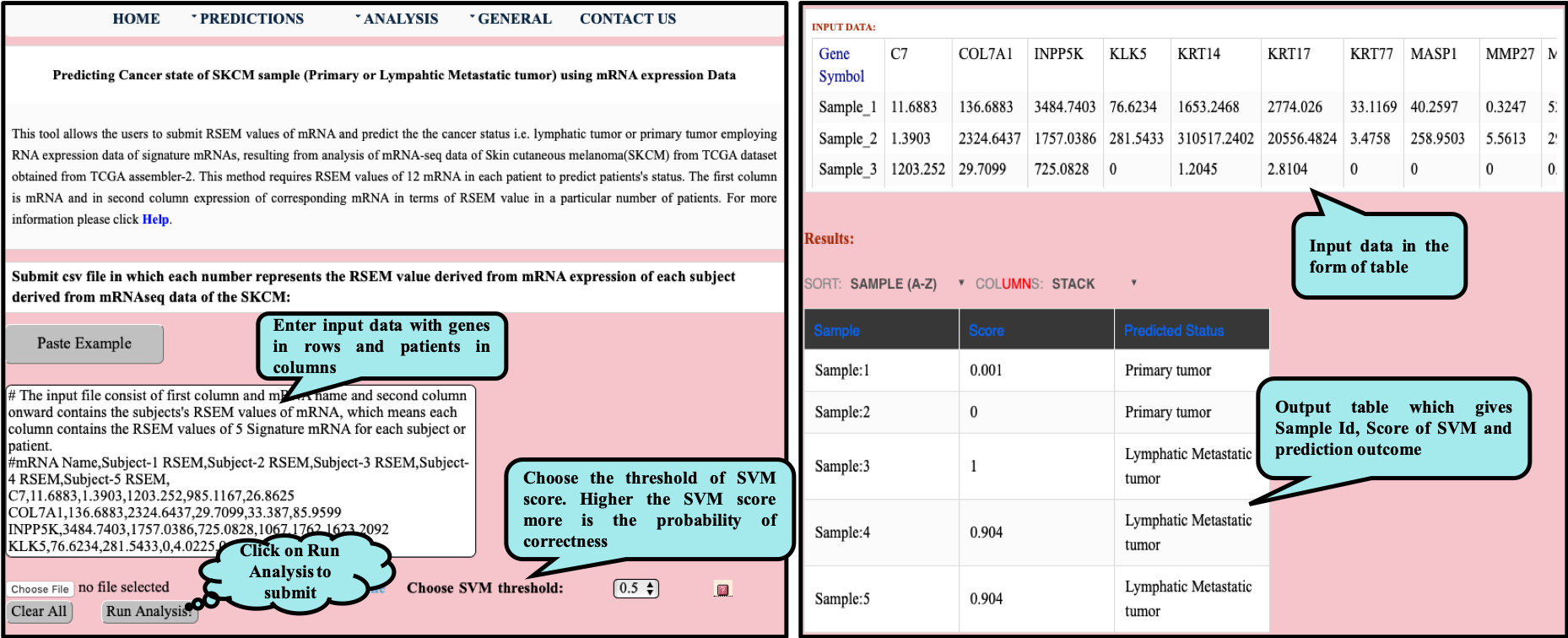
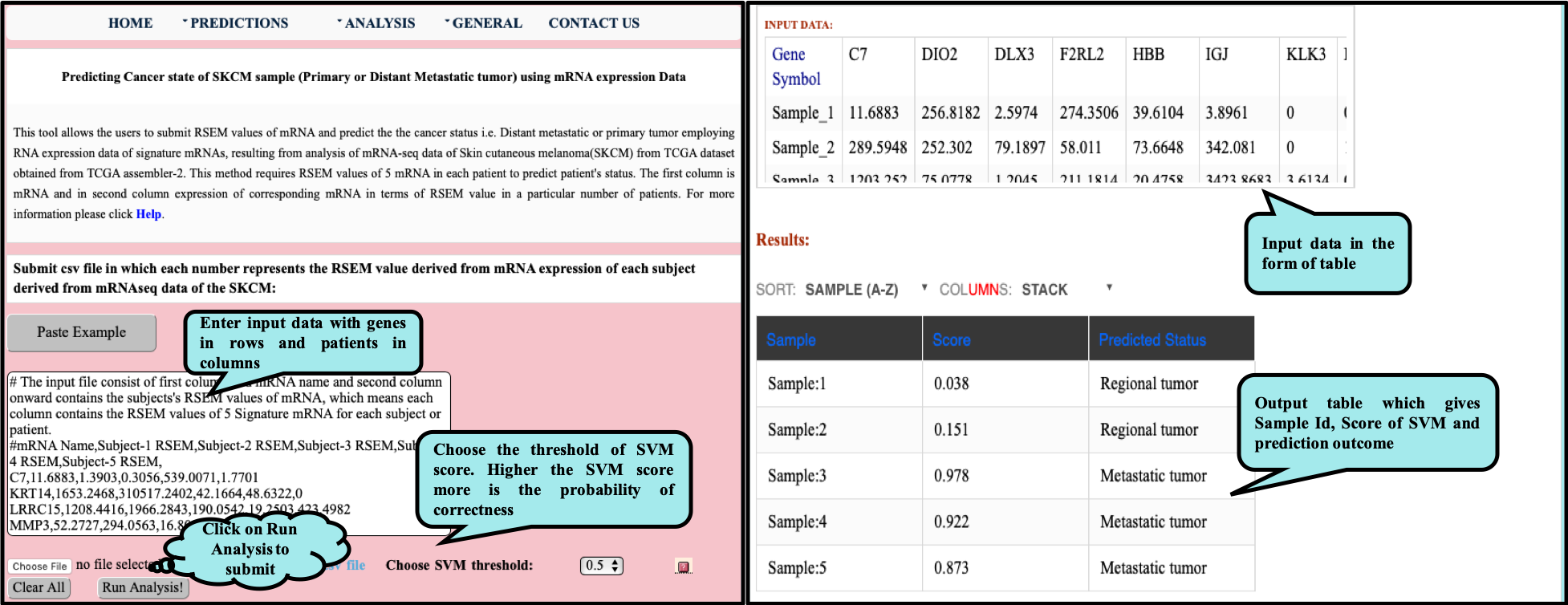
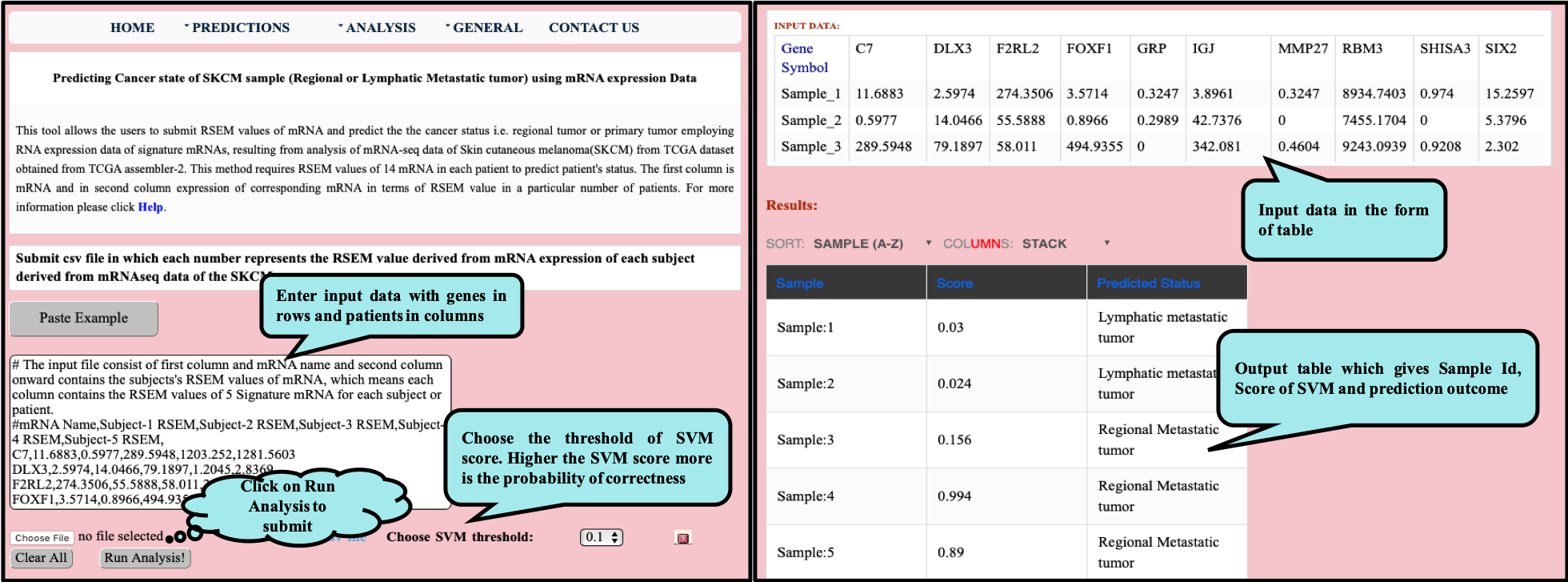
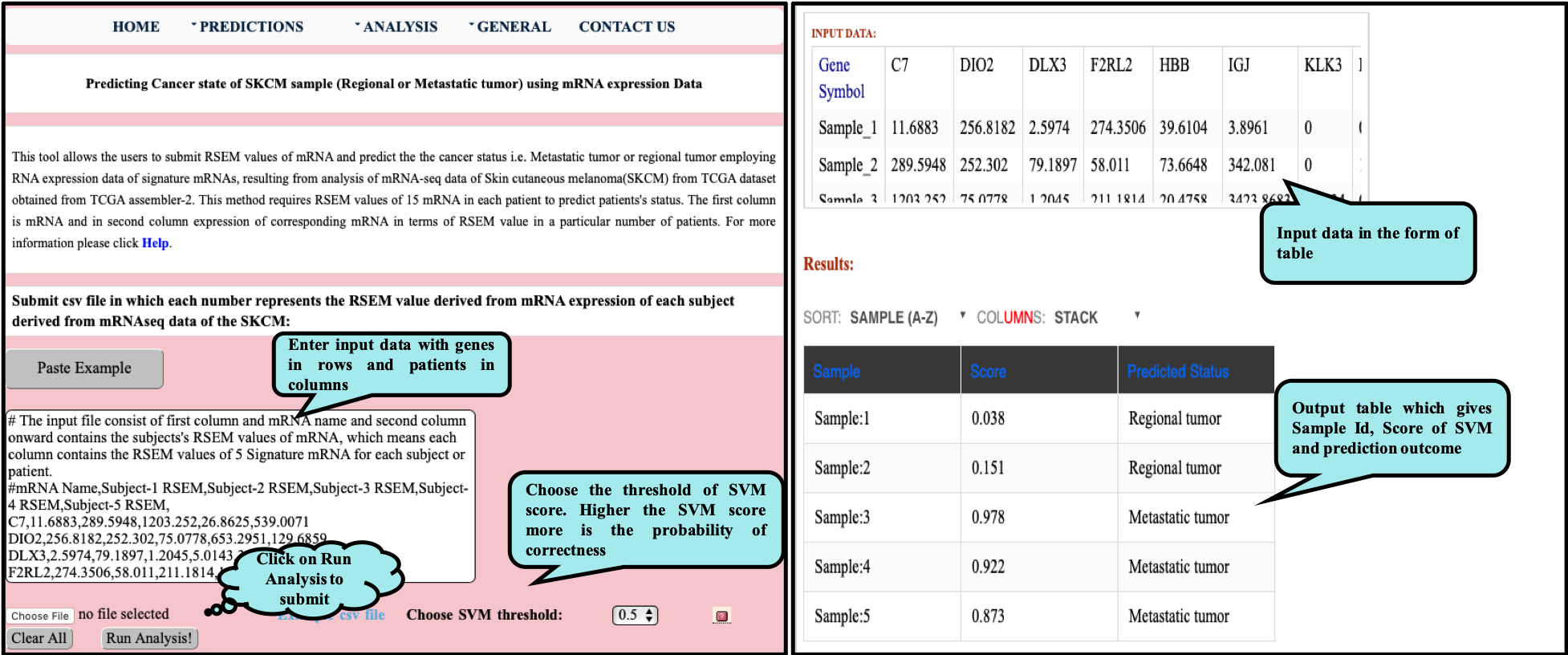
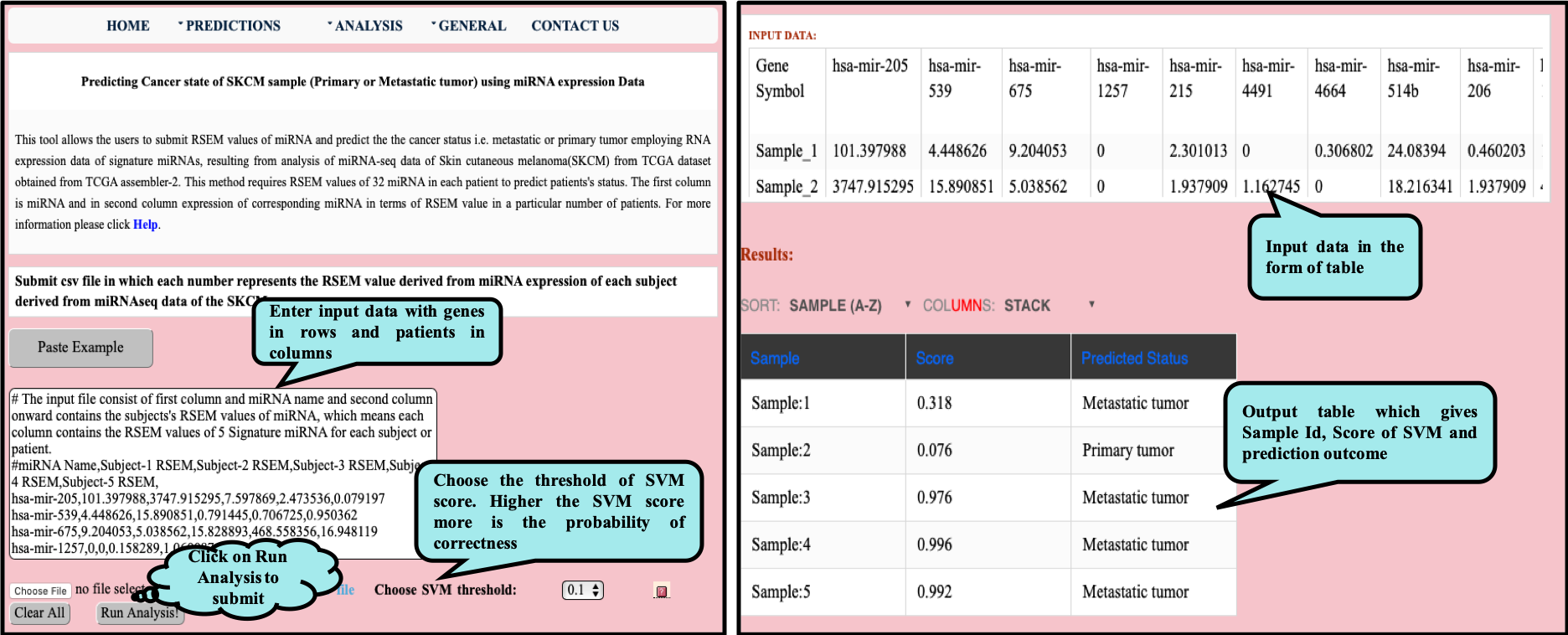
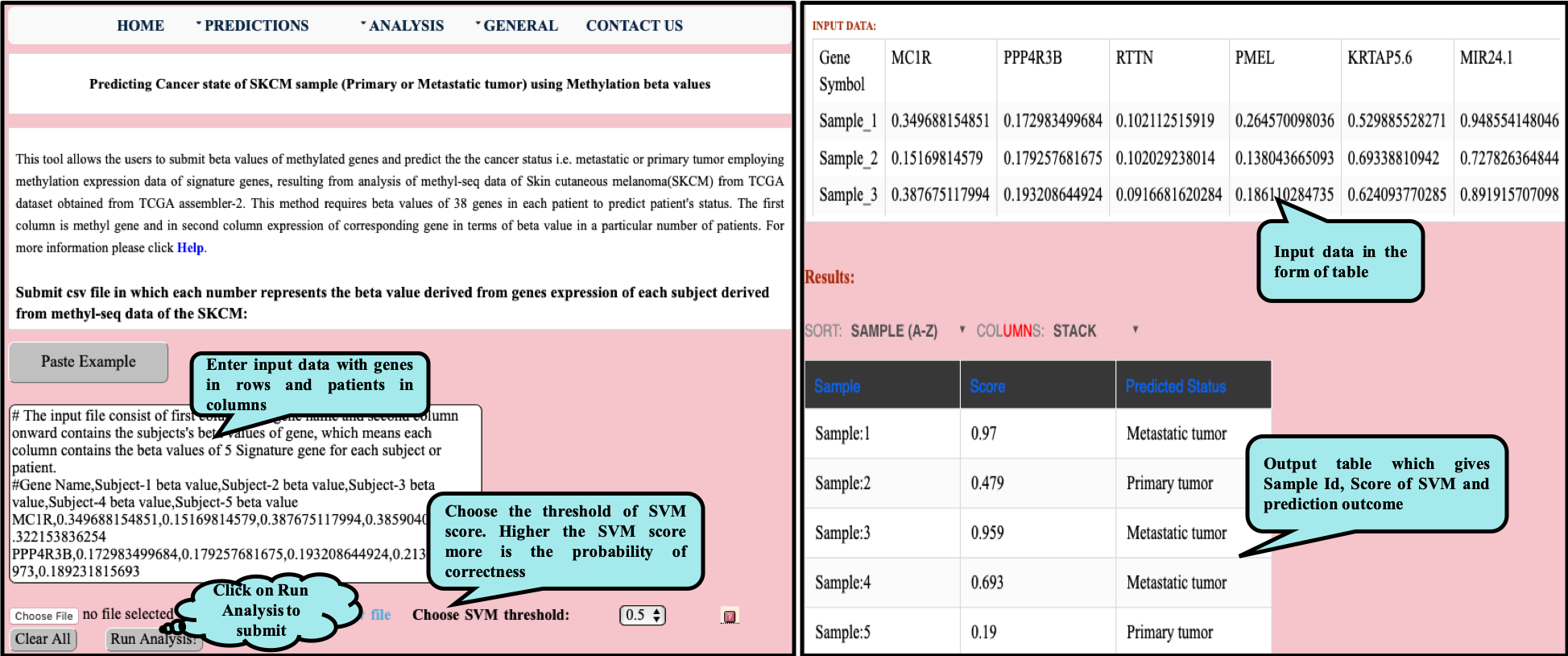
CancerSPP is a web-bench for skin cutaneous melanoma (SKCM) progression prediction. It is established for the prediction and analysis of primary and metastatic tumor of SKCM using signature genes expression data (derived from RNA-seq, miRNA and methylation RSEM expression quantification values) . Further, prediction module is used to predict multiple states of metastatic samples from primary tumor samples. Analysis module allow the user to analyze RNA-seq expression profiles data in primary and metastatic states of SKCM.
| Predictions | Analysis |
|---|