CancerEnD - A comprehensive resource on enhancer information for TCGA cancer types
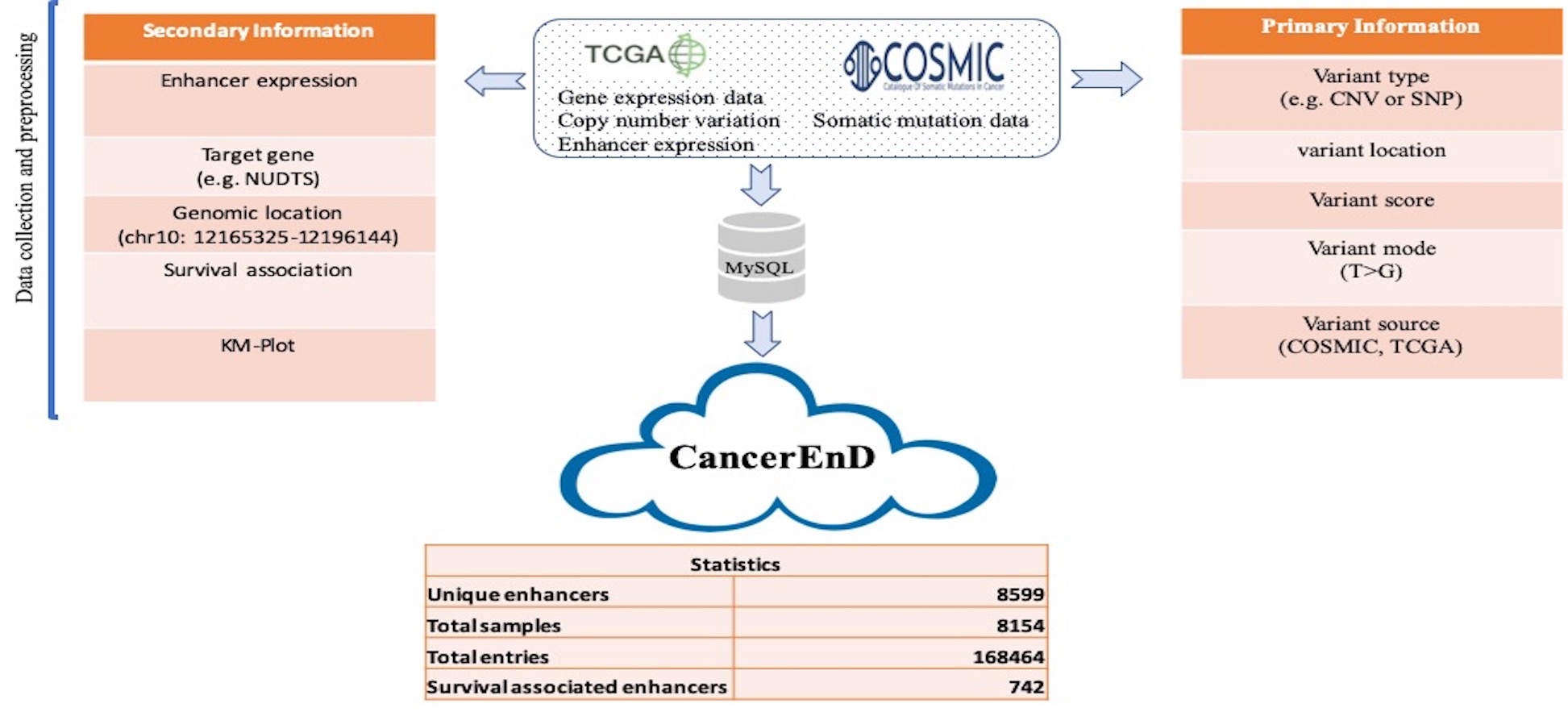
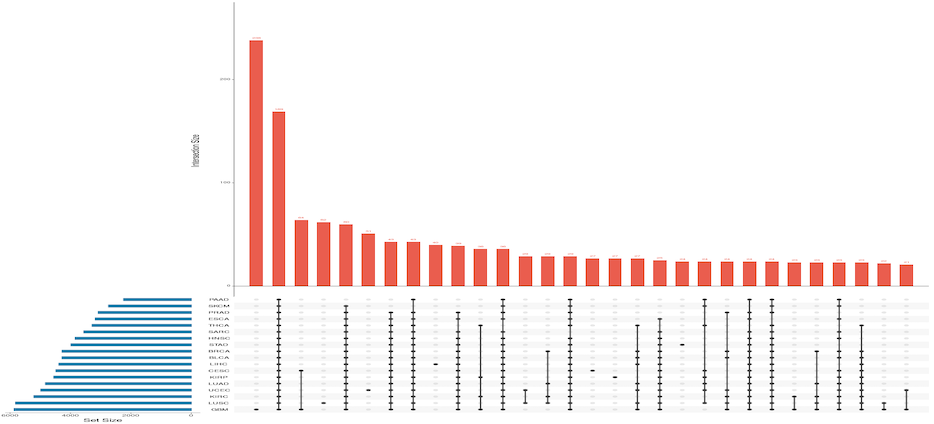
CancerEnD Genome-wide association studies have successfully identified thousands of genomic loci potentially associated with hundreds of complex traits in the past decade. Nevertheless, the fact that more than 90% of such disease-associated variants lie in non-coding DNA with unknown functional implications has been appealing for advanced analysis of plenty of genetic variants. Toward this goal, recent studies focusing on individual non-coding variants have revealed that complex diseases are often the consequences of erroneous interactions between enhancers and their target genes. CancerEnD is a database of Enhancers expression, their somatic mutation & CNV in 18 cancer types by extracting data from various large repositoies This database provides a user-friendly interface for browsing and searching, and it also allows users to download data freely. CancerEnD has the potential to become a helpful and important resource for researchers who aim to understand the molecular mechanisms of enhancers involved in complex diseases.
References (If you are using above please cite following articles)
- Kumar R., Lathwal A., Kumar V., Patiyal S., Raghav PK., and Raghava GPS (2020) CancerEnD: A database of cancer associated enhancers. Elsevier, Genomics 2020 May 1;S0888-7543(20)30264-0 (pmid:32360910)
- Kumar R et al., (2019) In Silico Analysis of Gene Expression Change Associated With Copy Number of Enhancers in Pancreatic Adenocarcinoma. MDPI, International Journal of Molecular Sciences, DOI: 10.3390/ijms20143582 (pmid:31336658)