Q1. What information does CancerEnD provides?
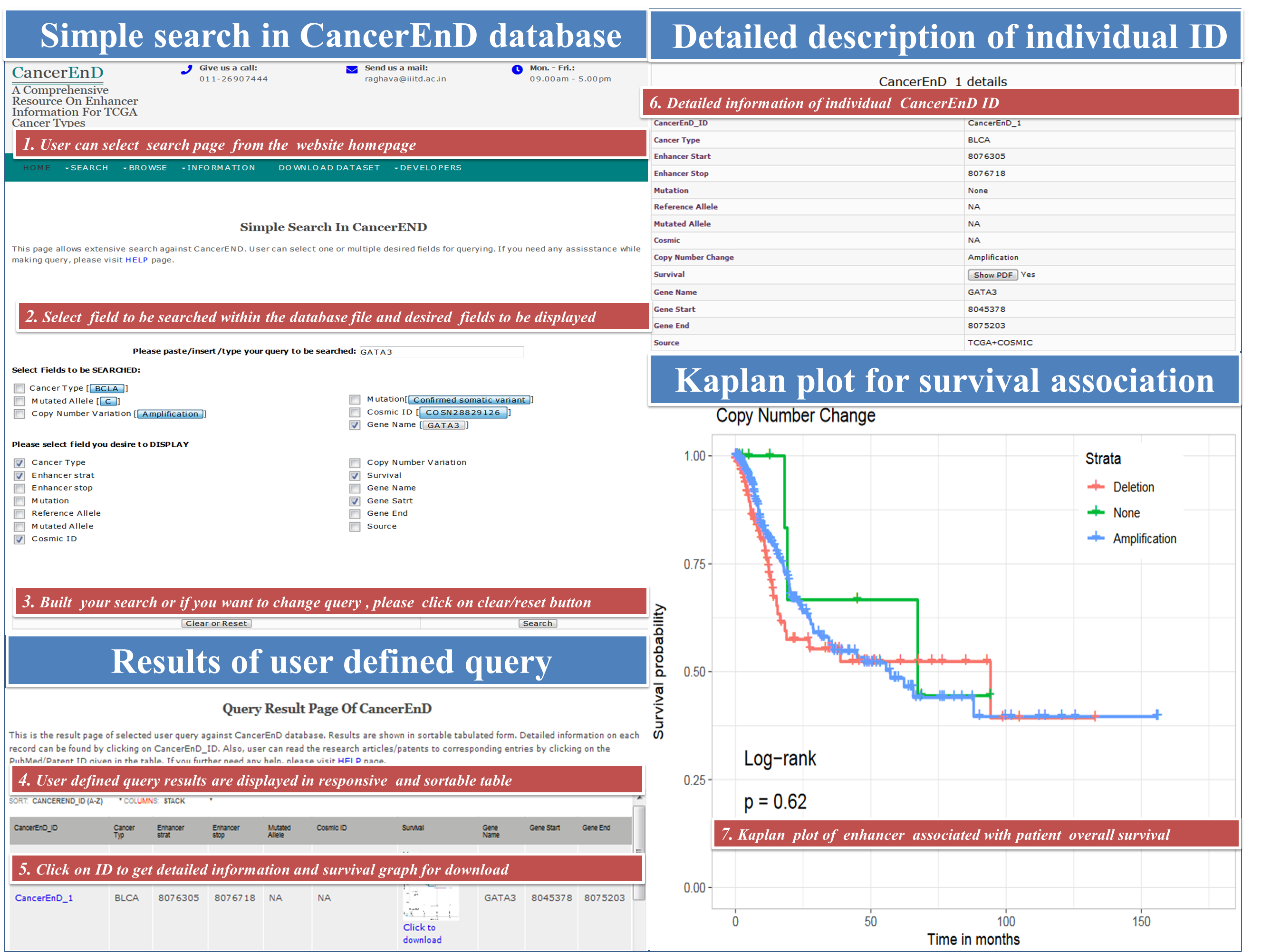
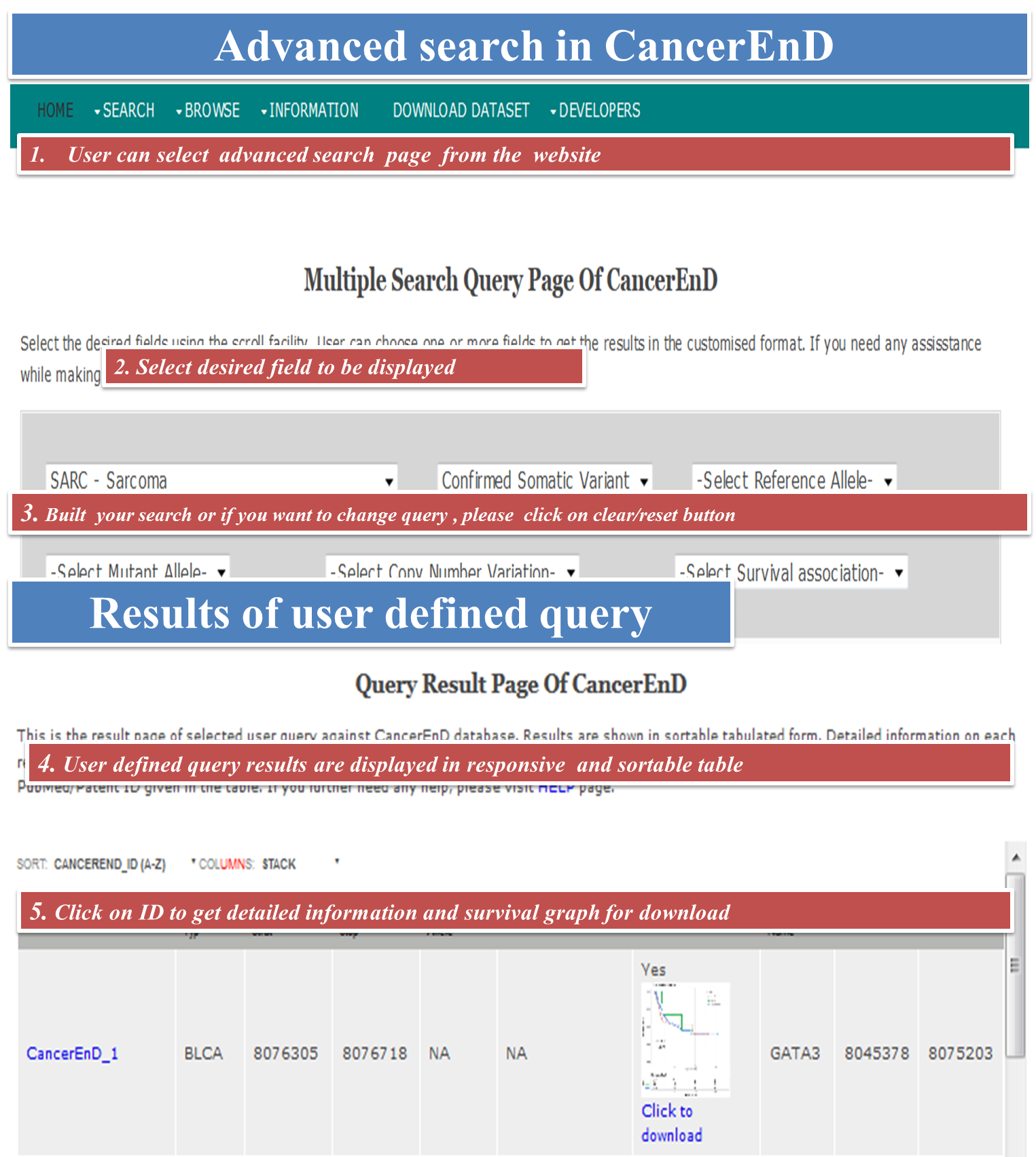
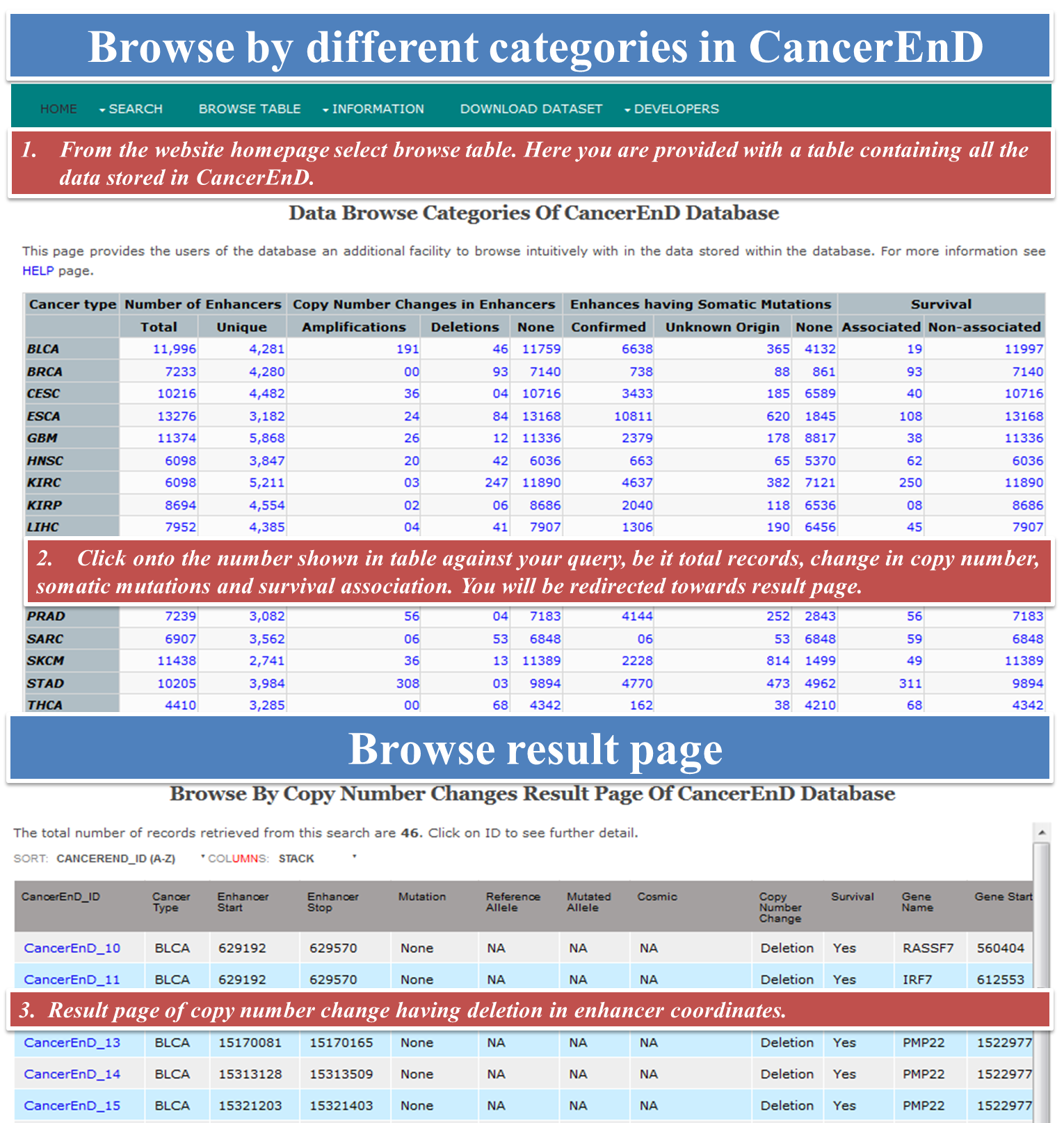
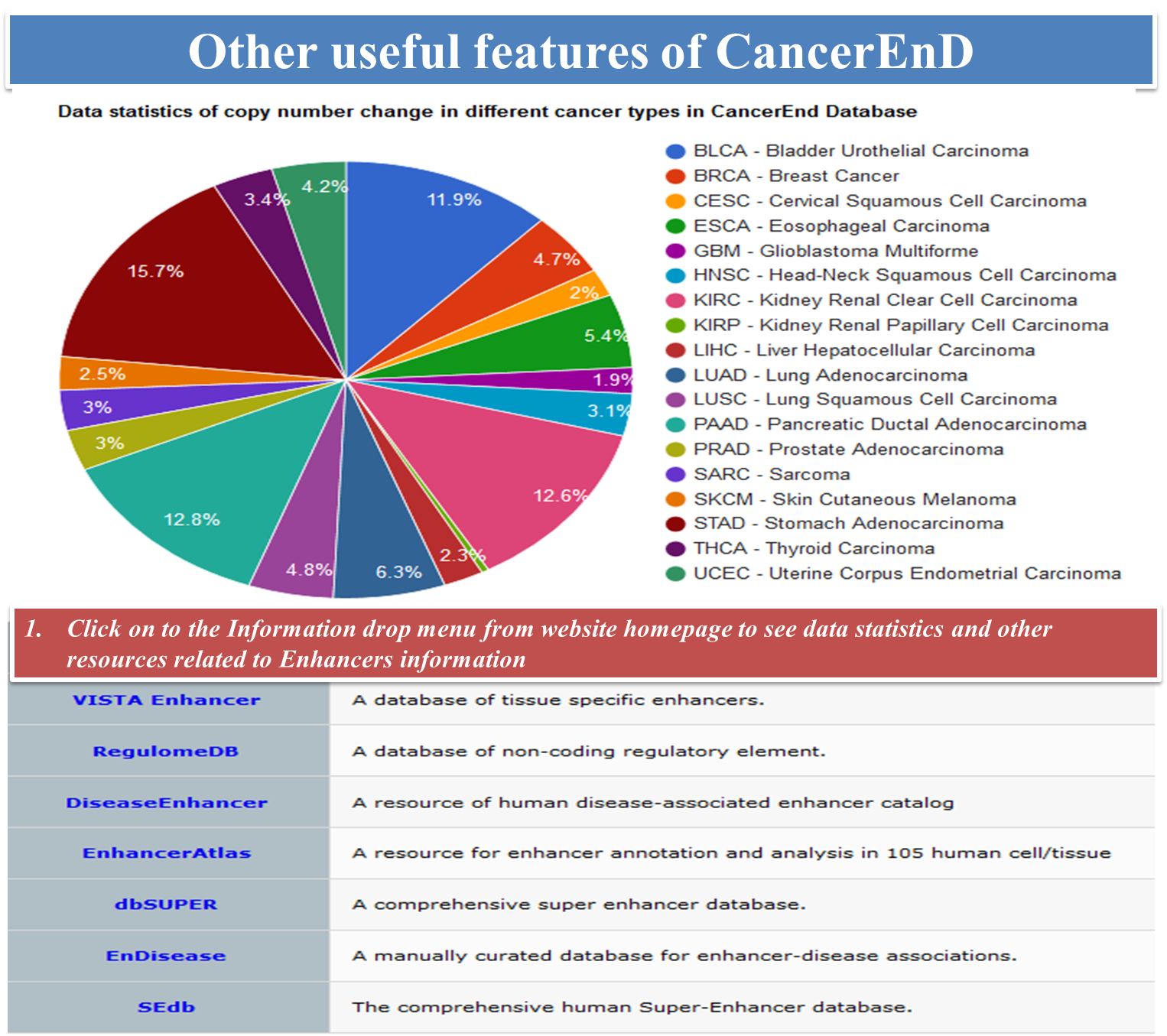
CancerEnD (Cancer En- Enhancer D- Database) is a repository of enhancer element, built by taking data from TCGA and COSMIC. It provides information on enhancer associated copy number change, linkage to patient survival, somatic mutation, enhancer expression across cancer patient wise and enhancer associated genes.
Q2. What can CancerEnD do?
With CancerEnD you can:
- look for common and unique enhancer for among each cancer type present in database.
- browse for different functional categories of enhancer like cancer type, copy number change, somatic mutation and survival.
Q3. Can the data stored in CancerEnD is downloadable?
User can download the entire data stored in CancerEnD database in three format viz. plain text, xlsx and in CSV format.
|
|