 |
| Home | submission | Help | Supplementary | Epitope Prediction | Developers |
BTXpred Supplementary Page
Datasets
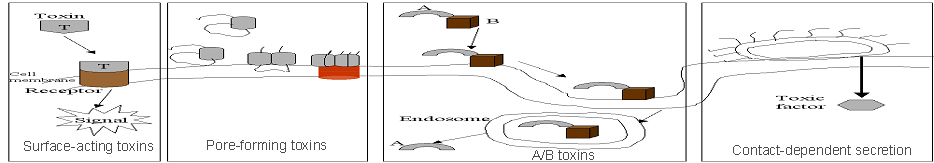
We searched bacterial toxins in Swiss-Prot( Bairoch and Apweiler, 2000) database using keyword toxin and (using NOT toxin). We examine each protein manually obtained from our query sequence in order to eliminate non bacterial toxins. Finally we obtained 185 (99 exotoxins and 86 endotoxins) proteins. We examined full entry (Swiss-Prot) of these toxins and classify them based on their molecular targets.
The non-toxins were obtained from Swiss-Prot by combined search using SRS. The search in the query from was performed along with "BUTNOT" option, using two information fields i) Comment with query word "function" , and ii) Comment using "toxin" as query word The retrived protein sequences were examined manually inorder to eliminate toxin proteins.The broad functions of the nontoxins can be viewed by clicking it here
The files can be downloaded:
The dataset for classificaton of bacterial toxins and non-toxin
The broad functions of the nontoxins can be viewed by clicking it here
The dataset for types of toxin based on release
- Endotoxin 86
Endotoxins are incorporated into cell wall and released into host tissues, when the bacteria die.
- Exotoxins 99
Exotoxins are soluble substance secreted into host tissues. These toxins have special affinity for particular tissues.
The dataset for functions of exotoxins
Endotoxins are incorporated into cell wall and released into host tissues, when the bacteria die.
Exotoxins are soluble substance secreted into host tissues. These toxins have special affinity for particular tissues.
The performance of various SVM modules on classification of bacterial toxins from non-toxins using amino acid composition, at threshold value of 0.1
Table S1: Various parameters were used to check the performance of the SVM module based on amino acid composition to classify bacterial toxins
|
RBF parameters |
Sensitivity |
Specificity |
Accuracy |
PPV |
MCC |
|
g=.1 c=100 j=1 |
0.8571 |
0.9357 |
0.8964 |
0.9386 |
0.8044 |
|
g=.1 c=1000 j=1 |
0.8571 |
0.9000 |
0.8786 |
0.9126 |
0.7693 |
|
g=.1 c=5000 j=1 |
0.8571 |
0.9143 |
0.8857 |
0.9214 |
0.7824 |
|
g=1 c=10 j=1 |
0.8571 |
0.9357 |
0.8964 |
0.9386 |
0.8044 |
|
g=1 c=100 j=1 |
0.8643 |
0.9143 |
0.8893 |
0.9238 |
0.7910 |
|
g=1 c=1000 j=1 |
0.8571 |
0.9571 |
0.9071 |
0.9576 |
0.8250 |
|
g=1 c=5000
j=1 |
0.9214 |
1.0000 |
0.9607 |
1.0000 |
0.9293 |
|
g=5 c=10 j=1 |
0.8643 |
0.9429 |
0.9036 |
0.9463 |
0.8187 |
|
g=5 c=100 j=1 |
0.8714 |
0.9714 |
0.9214 |
0.9711 |
0.8531 |
|
g=5 c=1000 j=1 |
0.9000 |
0.9714 |
0.9357 |
0.9729 |
0.8801 |
|
g=5 c=5000 j=1 |
0.9000 |
0.9714 |
0.9357 |
0.9729 |
0.8801 |
|
g=10 c=1 j=1 |
0.8571 |
0.9357 |
0.8964 |
0.9386 |
0.8044 |
|
g=10 c=10 j=1 |
0.8786 |
0.9500 |
0.9143 |
0.9523 |
0.8394 |
|
g=10 c=100 j=1 |
0.8857 |
0.9714 |
0.9286 |
0.9726 |
0.8663 |
|
g=10 c=1000 j=1 |
0.9000 |
0.9714 |
0.9357 |
0.9729 |
0.8801 |
|
g=10 c=5000 j=1 |
0.9000 |
0.9714 |
0.9357 |
0.9729 |
0.8801 |
The performance of various SVM modules on classification of bacterial toxins from non-toxins using dipeptide composition, at threshold value of 0.1
Table S2: Various parameters were used to check the performance of the SVM module based on dipeptide composition to classify bacterial toxins
|
RBF parameters |
Sensitivity |
Specificity |
Accuracy |
PPV |
MCC |
|
g=.1 c=1000 j=1 |
0.8571 |
0.9857 |
0.9214 |
0.9845 |
0.8547 |
|
g=.1 c=5000 j=1 |
0.8500 |
0.9643 |
0.9071 |
0.9602 |
0.8246 |
|
g=1 c=100 j=1 |
0.8571 |
0.9857 |
0.9214 |
0.9845 |
0.8547 |
|
g=1 c=1000 j=1 |
0.8500 |
0.9643 |
0.9071 |
0.9602 |
0.8246 |
|
g=1 c=5000 j=1 |
0.8500 |
0.9643 |
0.9071 |
0.9602 |
0.8246 |
|
g=5 c=10 j=1 |
0.8643 |
0.9714 |
0.9179 |
0.9711 |
0.8469 |
|
g=5 c=100 j=1 |
0.8500 |
0.9643 |
0.9071 |
0.9602 |
0.8246 |
|
g=5 c=1000 j=1 |
0.8500
|
0.9643 |
0.9071 |
0.9602 |
0.8246 |
|
g=5 c=5000 j=1 |
0.8500 |
0.9643 |
0.9071 |
0.9602 |
0.8246 |
|
g=10 c=10
j=1 |
0.8643 |
0.9857 |
0.9250 |
0.9849 |
0.8612 |
|
g=10 c=100 j=1 |
0.8571 |
0.9643 |
0.9107 |
0.9607 |
0.8305 |
|
g=10 c=1000 j=1 |
0.8571 |
0.9643 |
0.9107 |
0.9607 |
0.8305 |
|
g=10 c=5000 j=1 |
0.8571 |
0.9643 |
0.9107 |
0.9607 |
0.8305 |
|
g=25 c=10 j=1 |
0.8643 |
0.9786 |
0.9214 |
0.9782 |
0.8542 |
|
g=25 c=100 j=1 |
0.8571 |
0.9571 |
0.9071 |
0.9541 |
0.8235 |
|
g=25 c=1000 j=1 |
0.8571 |
0.9571 |
0.9071 |
0.9541 |
0.8235 |
|
g=25 c=5000 j=1 |
0.8571 |
0.9571 |
0.9071 |
0.9541 |
0.8235 |
The
performance of various SVM modules on classification of types of bacterial
toxins (Exotoxins or endotoxins) using features such as amino acid at threshold
value of 0.000
Table S3: Various parameters were used to check the performance of the SVM module based on amino composition to classify types of bacterial toxins, exotoxin and endotoxin
|
RBF parameters |
Sensitivity |
Specificity |
Accuracy |
PPV |
MCC |
|
g=1 c=10 j=1 |
0.6286 |
0.9000 |
0.7643 |
0.8635 |
0.5522 |
|
g=1 c=100 j=1 |
0.7286 |
0.8714 |
0.8000 |
0.8586 |
0.6159 |
|
g=5 c=100 j=1 |
0.7857 |
0.8857 |
0.8357 |
0.8767 |
0.6798 |
|
g=25 c=10 j=1 |
0.8571 |
0.9143 |
0.8857 |
0.9080 |
0.7783 |
|
g=25 c=100 j=1 |
0.8286 |
0.9143 |
0.8714 |
0.9064 |
0.7528 |
|
g=50 c=10 j=1 |
0.8429 |
0.9143 |
0.8786 |
0.9055 |
0.7629 |
|
g=50 c=100 j=1 |
0.8571 |
0.9143 |
0.8857 |
0.9092 |
0.7788 |
|
g=100 c=10 j=1 |
0.9000 |
0.9143 |
0.9071 |
0.9158 |
0.8211 |
|
g=100 c=100 j=1 |
0.8857
|
0.9143 |
0.9000 |
0.9136 |
0.8089 |
|
g=150 c=10 j=1 |
0.9286 |
0.9143 |
0.9214 |
0.9199 |
0.8501 |
|
g=150 c=100 j=1 |
0.9286 |
0.9143 |
0.9214 |
0.9199 |
0.8501 |
|
g=200 c=10 j=1 |
0.9571 |
0.9143 |
0.9357 |
0.9227 |
0.8760 |
|
g=200 c=100 j=1 |
0.9571 |
0.9143 |
0.9357 |
0.9227 |
0.8760 |
|
g=250 c=10
j=1 |
1.0000 |
0.9143 |
0.9571 |
0.9264 |
0.9203 |
|
g=250 c=100 j=1 |
1.0000 |
0.9143 |
0.9571 |
0.9264 |
0.9203 |
|
g=300 c=10 j=1 |
1.0000 |
0.9000 |
0.9500 |
0.9172 |
0.9085 |
The performance of various SVM
modules on classification of types of bacterial toxins (Exotoxins or
endotoxins) using features such as dipeptide composition at threshold value
of 0.000
Table S4: Various
parameters were used to check the performance of the SVM module based on
dipeptide composition to classify types of bacterial toxins, exotoxins and
endotoxins
|
RBF parameters |
Sensitivity |
Specificity |
Accuracy |
PPV |
MCC |
|
g=1 c=100 j=1 |
0.7857 |
0.9143 |
0.8500 |
0.9065 |
0.7084 |
|
g=1 c=1000 j=1 |
0.8000 |
0.9000 |
0.8500 |
0.9105 |
0.7250 |
|
g=10 c=100 j=1 |
0.8571 |
0.9000 |
0.8786 |
0.9105 |
0.7717 |
|
g=10 c=1000 j=1 |
0.8571 |
0.9000 |
0.8786 |
0.9105 |
0.7717 |
|
g=50 c=10 j=1 |
0.8857 |
0.9000 |
0.8929 |
0.9090 |
0.7929 |
|
g=50 c=100 j=1 |
0.8857 |
0.9000 |
0.8929 |
0.9090 |
0.7929 |
|
g=50
c=1000 j=1 |
0.8857
|
0.9000 |
0.8929 |
0.9090 |
0.7929 |
|
g=100 c=10 j=1 |
0.9429 |
0.9143 |
0.9206 |
0.9206 |
0.8596 |
|
g=100
c=100 j=1 |
0.9429 |
0.9143 |
0.9286 |
0.9206 |
0.8596 |
|
g=100 c=1000 j=1 |
0.9429 |
0.9286 |
0.9286 |
0.9206 |
0.8596 |
|
g=150 c=10 j=1 |
0.9429 |
0.9000 |
0.9214 |
0.9092 |
0.8456 |
|
g=150 c=100 j=1 |
0.9429 |
0.9000 |
0.9214 |
0.9092 |
0.8456 |
|
g=150 c=1000 j=1 |
0.9429 |
0.9000 |
0.9214 |
0.9092 |
0.8456 |
|
g=200 c=1 j=1 |
0.9000 |
0.8714 |
0.8857 |
0.8777 |
0.7739 |
|
g=200 c=10 j=1 |
0.9429 |
0.8857 |
0.9143 |
0.8992 |
0.8322 |
|
g=200 c=100 j=1 |
0.9429 |
0.8857 |
0.9143 |
0.8992 |
0.8322 |
Table S5: Showing summary of the individual entries based on SWISS-PROT accession number. PDB codes are also included and are linked to PDB database for structural information
|
Types of
Toxin |
Description |
Sub-types |
SWISS-PROT Number |
PDB codes |
|
|
E X O T O X I N |
Entero toxin |
Enterotoxins act on tissues of the gut and are further divided into three subclasses. |
Activate
adenylate cyclase |
P01555, P13810,P43528 P43530,P06717 |
1XTC , 1TII , 1HTL, 1LT3, 1LTT, 1LT4 , 1LTA 1LTA, 1LTG , 1LTI, 1LTS |
|
Activate
guanylate cyclase |
P01559, Q47185, P07965, P07965,P01560, P74977, O50319, P22542 |
||||
Foodpoisioning |
P01558, P01553,P34071, P23313,P0A0L2, P01552, P20723,P12993, O85382, P0A0M0 |
1CQV , 1I4P , 1I4Q ,1I4R , 1I4X , 1SE2 ,1STE , 1UNS 1DYQ.1ESF, 1I4G, 1I4H, 1LO5, 1SXT,1D5M,1D5X, 1D5Z, 1D6E,1SBB, 1SE3, 1SE4,1SEB, 1ENF, 1EWC |
|||
|
Neuro toxin |
Neurotoxins act on tissues of the nervous system |
|
P10845, Q45894,P10844, P18640P19321, Q00496 P30995, P30996 Q60393 |
||
|
Cyto toxin |
Cytotoxins act on general tissues |
Thiol-activating |
P19995, P13128, P23564, Q53957Q54114, P21131 |
|
|
|
Vacuolating
cytotoxin |
Q48247, Q48245Q48253, Q48258Q9ZKW5,P55981 |
|
|||
|
Macrophage
cytotoxin |
P55129, P15377P55131 |
|
|||
Hemolysin |
P55870, Q08675,P09983, P28031,P19249, P19250 P28029, Q08677,P08715, P16466,P15320, Q06803 P09545, Q54316,P14711, P28030,P23182, P01506 P31714, P0A077,Q07227, Q00951,Q44066, P77335 P16535, P55116,P55117, P55118,P16462, P15310 Q9RF12, P20419,Q46150, P09978,P19247, P06200 P31715 |
|
|||
E
N DO T O X I N |
Endotoxins are incorporated into cell wall and released into host tissues, when the bacteria die. |
Insecticidal toxin |
P21256, Q45730,Q45754, Q45710,Q45729, Q45882 O05102, Q45358,P57091, P57092,O32307, O86170 P02965, P06578,P05068, Q03744,Q03748, P96315 Q9S515, P05517,Q45739, Q45774,Q9ZAZ5,O85805 P05518, P56953,P19415, Q45747,Q57458, Q03745 Q03746,O66377,Q45746,Q9ZAZ6,Q45748, Q45718 Q45752,Q45709,O87404,Q9XDL1,Q45738, Q45716 Q45715, O32321,P56956, P56957,O87905, O87906 Q9X597,Q9S597,Q9X682, P21253,P21254, Q45743 Q9RMG3, P07130,P17969 Q06117, Q45744,P16480, P05519,Q45760, Q45753 P56955, Q45712,Q45757, Q45758,Q03749, Q45707 Q45708, Q45704,Q45705, Q45706,Q99031, Q45733 O06014,Q9ZNL9,P09662, P05069,P94594, Q45790 Q04470, Q45723,O32322 |
|
|
Algorithm
Support vector machine
The SVM was implemented using freely downloadable software package SVM_light (Joachims, 1999). The software enables the user to define a number of parameters as well as to select from a choice of inbuilt kernel functions, including a radial basis function (RBF) and a polynomial kernel. Preliminary tests show that the radial basis function (RBF) kernel gives results better than other kernels. Therefore, in this work we use the RBF kernel for all the experiments. The input vectors used are amino acid composition (20 vectors) and dipeptide composition(400 vectors) of each protein sequence.
Protein features
Amino acid composition . Amino acid composition is the fraction of each amino acid in a protein.
Dipeptide composition.
Dipeptide composition was used to encapsulate the global information about each protein sequence, which gives a fixed pattern length of 400 (20 ´ 20). This representation encompassed the information about amino acid composition along local order of amino acid.
Hidden markov model
We generated HMM profiles of seven functional class of exotoxins using HMMER ((Eddy, 1998). Each functional class sequences were aligned in a multiple sequence alignment using CLUSTAL-W. A profile HMM was build with hmmbuild program for each functional class and later each profile was calibrated with hmmcalibrate program. We created our own HMM database by concating single HMM profile files. Hmmpfam program was used for searching a query sequence against the profile HMM databaes. We set an E-value threshold (e value>0.01) while predicting quality by a leave-one–out cross-validation.
Down load Hmm files
PSI-BLAST
A module of PSI-BLAST (Altschul et al., 1997) was designed in which query sequences in test dataset were searched against proteins in training dataset using PSI-BLAST. Three iterations of PSI-BLAST were carried out at a cut-off E-value of 0.01. The module could predict bacterial toxins toxins and types of toxins(exotoxin and endotoxin) and function of the exotoxins (activate adenylate cyclase, activate guanylate cyclase, food poisioning, neurotoxins, macrophage cytotoxin, vacuolating cytotoxin and thiol activated cytotoxin) depending upon the similarity of the query protein to the protein in the dataset.
Performance measures
Five-fold cross-validation
The performance modules constructed in this study for discriminating bacterial toxins, non-toxins and exotoxins and endotoxins were evaluated using a 5-fold cross-validation technique. In the 5-fold cross-validation, the relevant dataset was randomly divided into five sets. The training and testing was carried out five times, each time using one distinct set for testing and the remaining four sets for training. Five threshold-dependent parameters - sensitivity, specificity, accuracy, PPV and Matthew’s correlation coefficient (MCC), were used for discriminating bacterial toxins and non-toxins and also in classification of bacterial toxins into exotoxins and endotoxins.
Leave-one out cross-validation
We examined the prediction quality of functional classification of exotoxins by a leave-one-out cross-validation. During the process the leave-one out cross-validation, the training (46 sequences)and testing (one sequence) datasets are open, and a protein will turn move from one to the other.HMM profile database was made 46 times using 46 sequences at a time as testing set and using one protein sequence as testing set. Similarly PSI-Blast dataset was made 46 times leaving one protein sequence, which was used as testing set.
ClustalX (Multiple sequence analysis)
MUltilpe sequence analysis of seven different functions of exotoxins are available.
click here for clustalx sequence analysis.
PDB codes of bacterial toxins are available.
click here for viewing PDB codes of bacterial toxins.
[ Contact ] [ BIC, IMTECH ]