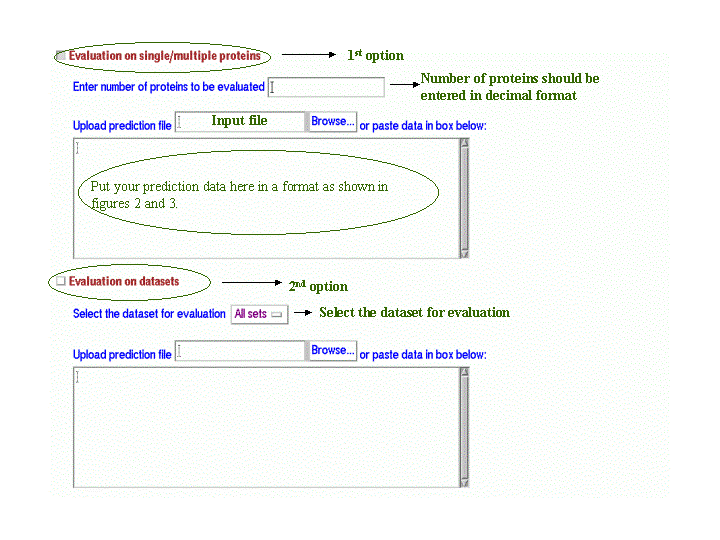
For evaluation of beta-turn prediction method, there are two options: (marked as 1st option and 2nd option in figure below) Either you can do evaluation on single/multiple proteins or on subsets/complete dataset. As shown in figure below, there are checkboxes for these two options and you can tick either of them as per your requirement.
For evaluation on single/multiple proteins, you have to do following steps:
- Enter the number of proteins for which you have to do evaluation. The number of proteins should be entered in decimal format. For example, for evaluation of single protein, you can write 1 or if the number of proteins for evaluation are more than one, say seven, you can enter 7.
- Enter the input prediction data which can be pasted in the textarea provided or can be uploaded through file.
The input prediction data should be in the following format:

Figure 2
The first line ">1ah7" is the sign ">" followed by PDB code. The second line is the amino acid sequence in a single line without any line break. The third line is the turn/nonturn predictions of correponding residues. The predicted turns should be written as small "t" and predicted non-turns should be written as small "n". The predicted turn/nonturns should also be a single line without any line break.
The input prediction data for multiple proteins should look like this:
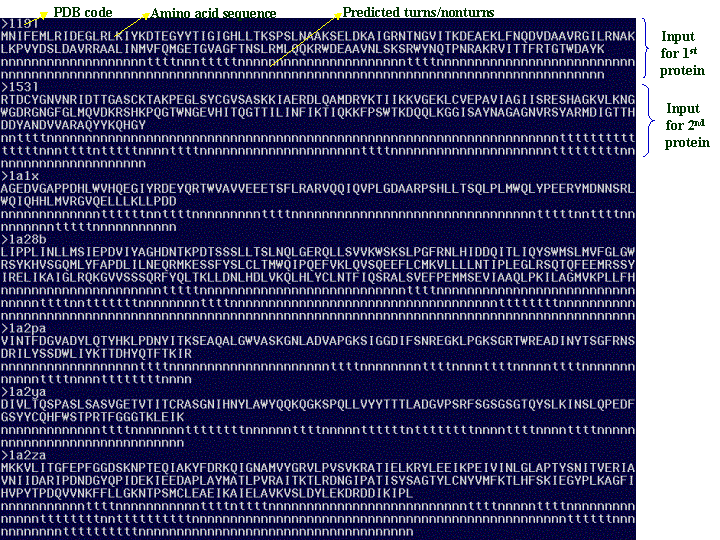 Figure 3
Figure 3
As shown in the above figure, there are 7 proteins and for each protein there are 3 lines: >PDB code, amino acid sequence and predicted turns/nonturns.
For evaluation on datasets, you have to do following steps:
- Select the dataset(setsI-VII or all sets) for which you have to do evaluation. You can download the respective dataset at this link. The datasets have protein sequences in fasta format.
- Enter the input prediction data which can be pasted in the textarea provided or can be uploaded through file. The format of input prediction data is same as the format for multiple proteins (Figure 3)
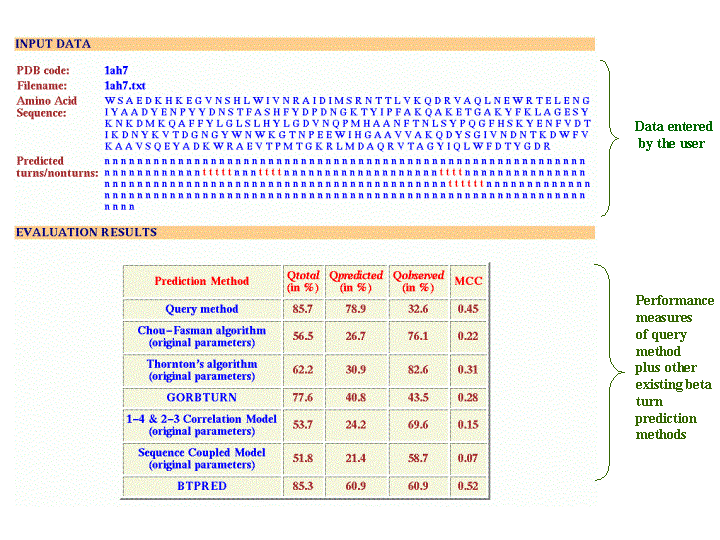
The output is in a tabular format that shows the performance measures(Qtotal, Qpred., Qobs. and MCC) of query method (for which the evaluation is carried out) along with the performance of other existing beta turn prediction methods such as Chou-Fasman algorithm, Thornton's algorithm, GORBTURN, 1-4 & 2-3 Correlation method, Sequence Coupled Model and BTPRED. For multiple proteins and datasets, the performance measures will be averaged over the number of proteins.
Output for a single protein
The ouput has 2 sections. The upper portion has input data as entered by the user. It contains the PDB code, filename if loaded, amino acid sequence of the protein and predicted turns/nonturns data. The lower portion has evaluation results in tabular form having the perormance measures Qtotal, Qpred., Qobs. and MCC. The results also display the performance of other existing methods on the query protein.
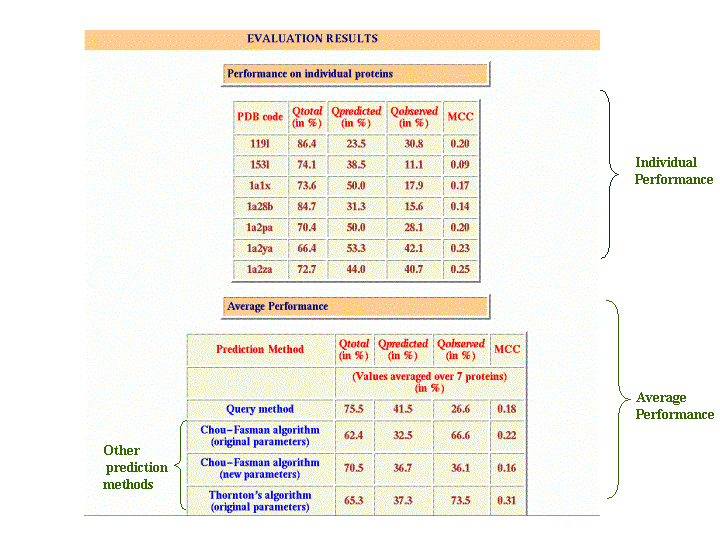
Output for multiple proteins/sets
The evaluation output of multiple proteins has 3 portions: input data, performance of query method on individual proteins and average performance(performance measures averaged over the number of proteins). Also displays the results for other existing beta-turn prediction methods.