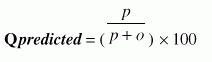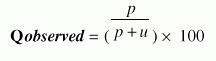| Department of Computational Biology |
| IMTECH, New Delhi, INDIA |
 |
 |
 |
 |
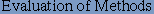 |
 |
 |
 |
 |
 |
 |
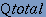 |
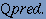 |
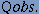 |
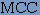 |
 |
 |
 |
| Here, four different parameters are used to measure the performance of BetaTPred2 as described by Shepherd et al. (1999). The predictive performance of a method is expressed by following four parameters: 1. Qtotal, the percentage of correctly classified residues, is defined as
where, p is the number of correctly classified beta-turn residues, n is the number of correctly classified non-beta-turn residues and t is the total number of residues in a protein. Qtotal, also known as 'prediction accuracy' may be defined simply as the total percentage of correct prediction. One difficulty with this measure is that it does not take into account disparities in the number of beta-turns(around 25%) and non-turns. Hence, it is possible to get a Qtotal score of about 75% by the trivial strategy of predicting all residues to be non-turn residues. Therefore, there is a risk of losing the information because of the dominance of non-turn residues. The Matthews Correlation Coefficient remedies this problem, which is defined as 2. MCC, the Matthews Correlation Coefficient, defined as
where, p is the number of correctly classified beta-turn residues, n is the number of correctly classified non-beta-turn residues, o is the number of non-beta-turn residues incorrectly classified as beta-turn residues and u is the number of beta-turn residues incorrectly classified as non-beta-turn residues. It is a measure that accounts for both over- and under-predictions. 3. Qpredicted, defined as
Qpredicted is the percentage of beta-turn predictions that are correct. Otherwise known as specificity, is the proportion of true negatives or the proportion of non-turn residues that have been correctly predicted as nonturns. 4. Qobserved, defined as
Qobserved is the percentage of observed beta-turns that are correctly predicted. Otherwise, known as sensitivity, is the proportion of true positives or the proportion of beta-turn residues that have been correctly predicted as beta-turns. Thus, the prediction accuracy is measured at residue level or accuracy is considered in terms of the percentage of individual amino acids predicted correctly. |