




| BetaTPred2: Prediction of ß-turns in proteins using neural networks and multiple alignment |
 |
 |
 |
 |
 |
 |
|
The purpose of BetaTPred2 server is to predict beta-turns in a protein from amino acid sequence. It uses the multiple alignment and secondary structure information for prediction. The prediction is done at amino acid level. About submission form Input SequenceThe input sequence for prediction is a one-letter code amino acid sequence in fasta or free format. The name of protein/query is optional. The sequence can be pasted in the text area provided or can be uploaded through a file in the format (fasta or free) specified in the form. 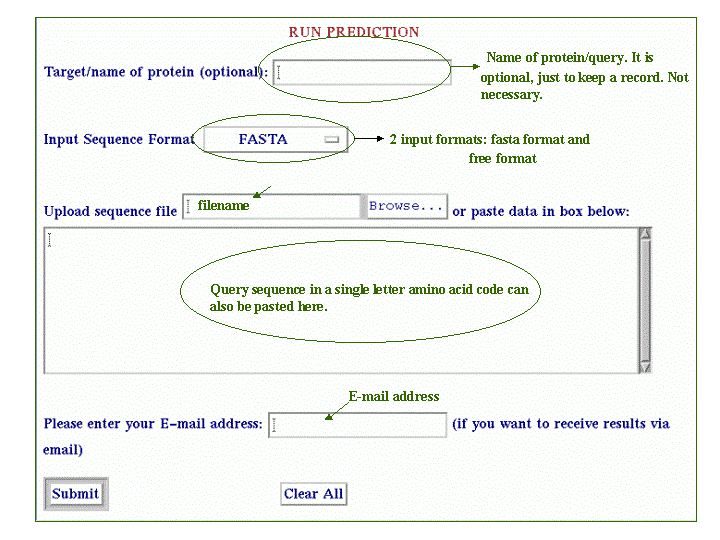 Shown below is the query sequence in fasta format pasted in the textarea. 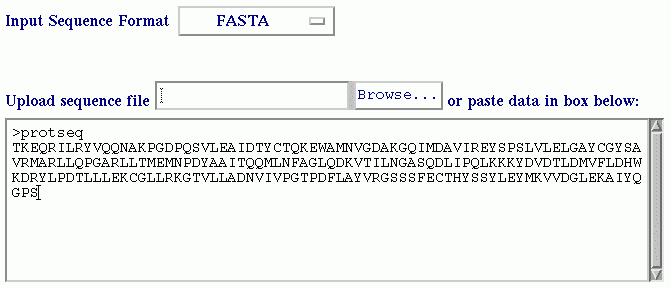 The prediction method uses the PSIPRED program to have the intermediate PSI-BLAST matrices and secondary structure. The whole process is bit time consuming so you can also get the results through E-mail. You can give the valid E-mail address in the E-mail box provided in the form. OutputThe output shows the input data as submitted by the user along with the prediction results. The prediction results consists of three rows, first row has the query sequence(submitted by the user), second row has the secondary structure predicted by PSIPRED. PSIPRED predicts 3 states: Helix denoted by 'H', Strand denoted by 'E' and Coil denoted by 'C'. The third row has the predicted turns/nonturns. The turns are denoted as small 't' and nonturns are denoted as small 'n'. 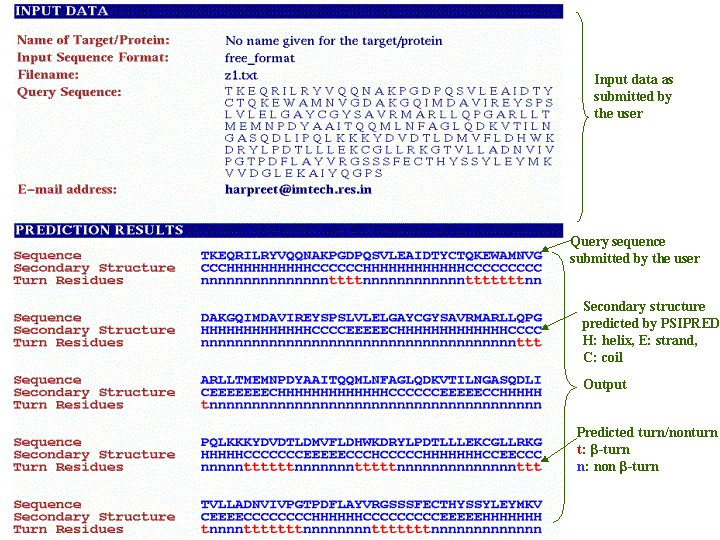
|