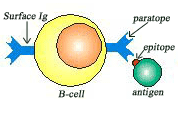
B-cell and epitope of antigen
|
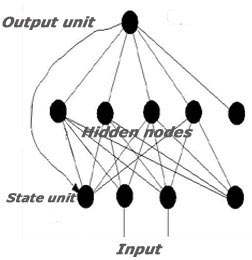
Recurrent neural network
|
|
|
|
The aim of ABCpred server is to predict linear
B cell epitope regions in an antigen sequence, using
artificial neural network. This server will assist in locating
epitope regions that are useful in selecting synthetic vaccine
candidates, disease diagonosis and also in allergy
research.
|
ABCpred has been trained on B cell epitopes
obtained from Bcipep database ( BCIPEP), and will therefore
presumbly have better performance for prediction of B cell epitope
of an antigen. This server can predict continuous (linear) B cell
epitopes. A linear B cell epitope is a short peptide that
cross-reacts with an antibody, which binds to a conformational
epitope.
|
|
Users can select window length of 10, 12, 14,
16 and 20 as predicted epitope length. It presents the
results in graphical and tabular frame. In case of graphical frame,
this server plot the epitopes in blue color along protein backbone
(black color), which assist the users in rapid visulaziation of
B-cell epitope on protein. The tabular output is in the form of a
table, which will provide the aminoacids length from N-terminal to
C-terminal in a protein predicted by the server.
|
|
The server is able to predict epitopes with
65.93% accuracy using recurrent neural network.
|
Please cite following paper if you are using
ABCpred server
| |






