|
|
|
Help Page |
| EGFRpred is a webserver for the classification of active and inactive anti-egfr compounds. It also allow the user to designing their analogs. Three major modules have been integrated to facilitate users. This page is designed for providing help on EGFRpred. Please click on topic for detail help. |
|
|
| IMPROTANT FINGERPRINTS |
| We have identified 126 fingerprints/substructures that play an important role in classification of active vs. inactive anti-cancer inhibitors. The fingerprints will help in better understanding of anti-cancer inhibitors. We encourage users to understand these 126 fingerprints, which will help them in all three modules: Draw structure, Virtual Screening and Analog Designing. Click here to view best 100 fingerprints of EGFR10 dataset in a tabular format. Click here to view best 100 fingerprints of EGFR100 dataset in a tabular format. Click here to view best 100 fingerprints of EGFR1000 dataset in a tabular format. |
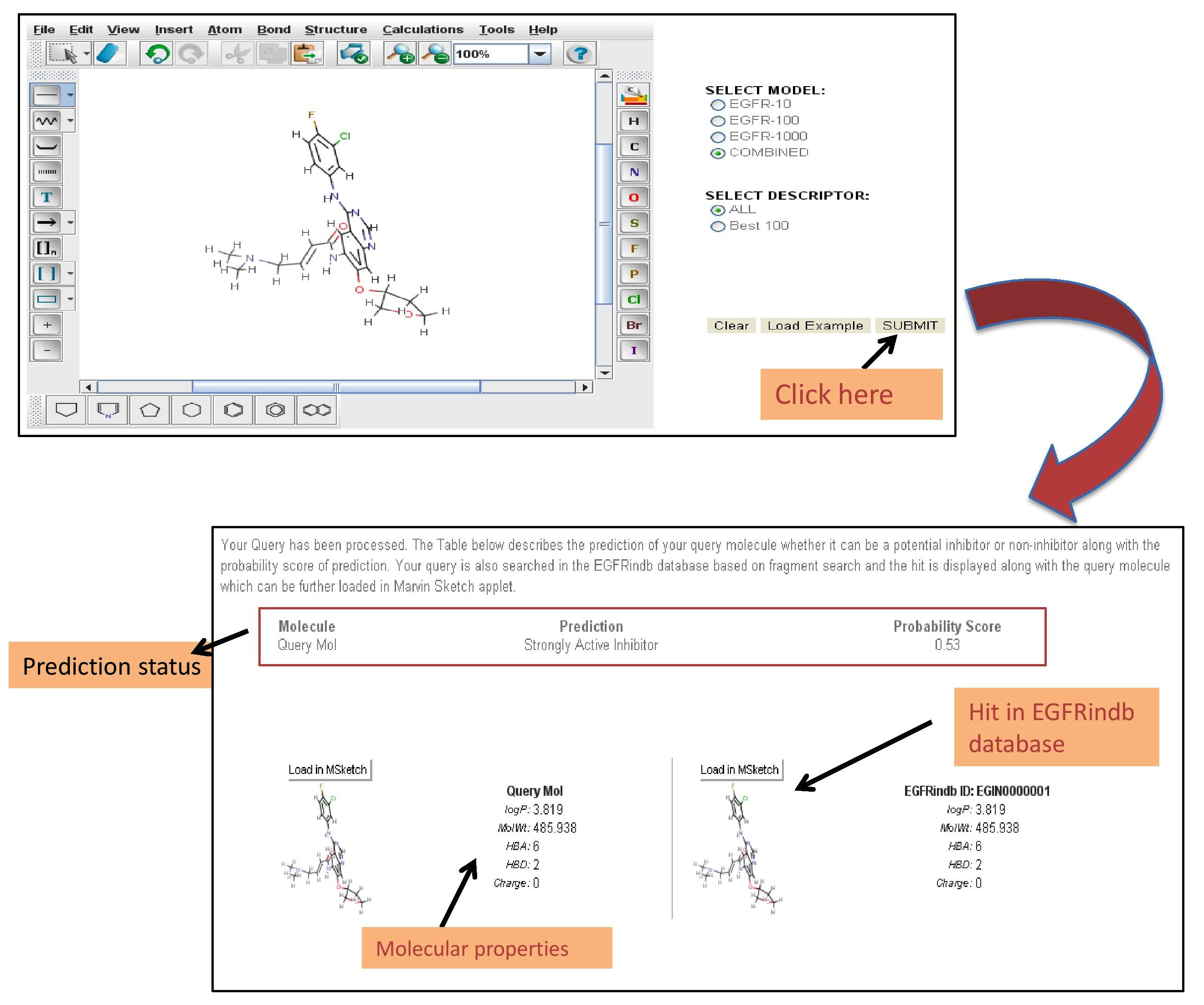
| DRAW A MOLECULE FOR PREDICTION: |
We have integrated Chemical Drawing java applet Marvin Sketch for drawing/loading a chemical structure. A user can also modify the structure using integrated options of Marvin Sketch applet like Acetylation/Amidation of the structure. We have also provided an example structure which can be loaded in the Marvin Sketch drawing box by clicking on "Load Example" button. When a user clicks on "submit" button, the backend processing is done by saving the chemical structure in sdf format and descriptors are calculated by PaDEL software which are then selected and finally prediction is done by applying machine learning method (Random forest) using weka package.
The figure below describes how to use "draw" module for drawing and prediction of chemical compounds for antiEGFR activity. |
 |
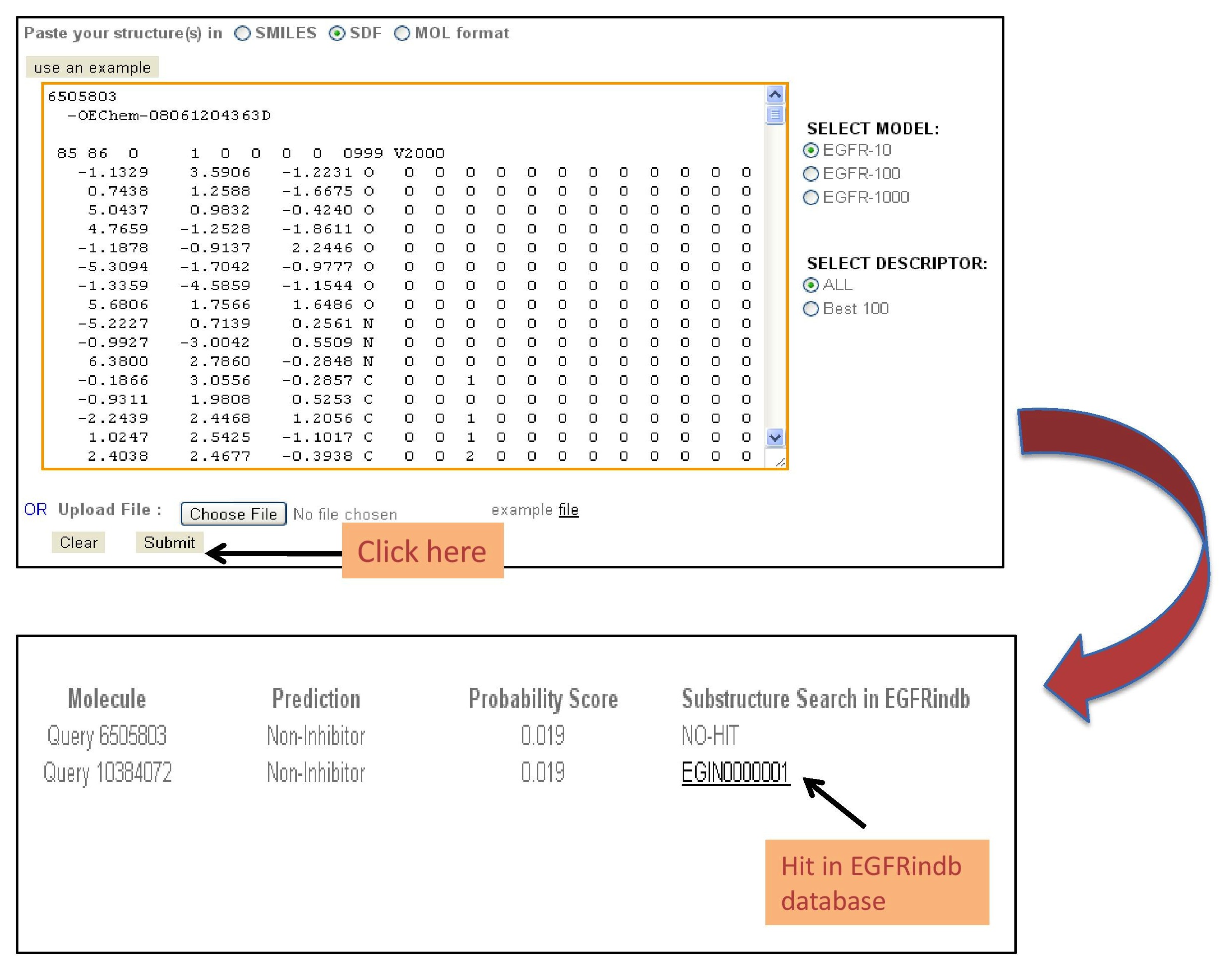
| VIRTUAL SCREENING OF A CHEMICAL LIBRARY: |
This module gives an option to submit multiple chemical molecules in the form of a chemical library and predict the anti-EGFR activity of each chemical in the library. There are 2 options to submit the chemical library. 1) A user can directly paste the multiple chemical molecules in the smiles/sdf format in the box provided. 2) A user can upload his/her chemical library file in the smiles or sdf format.
The figure given below describes the use of this module. |
 |
| DESIGN ANALOGS: |
This module gives an option to generate all the possible chemical analogs of a molecule based upon the combinations of user defined scaffolds, linkers and building blocks. The Server then predicts anti-EGFR activity of all the generated chemical analogs. In this way a user can identify new lead molecules for anti-EGFR activity.
The figure given below describes the use of this module. |
 |
|


