For the development of QSAR model, we have used total dataset of 98 molecules, which were taken from literature. Structure were drawn using ChemDraw software and then converted into 3D structure followed by energy minimization by openbabel software
Descriptor calculation:
In this step, we have calculated descriptors using different softwares, V-life, PaDEL, BlueDesc and CDK. V-life MDS (Molecular Design Suite) is a workbench for computer aided drug design (CADD) and molecule discovery. Through V-life software, we have calculated ~1002 descriptors including 1D,2D and 3D descriptors. PaDEL is an open source software that can calculates ~10 different types of binary fingerprints along with 2D and 3D descriptors. Another source is Chemistry Development Kit (CDK), a Java based open source library for structural chemo- and bioinformatics projects. by which we have computed 178 descriptors. BlueDesc, is another open source software based on CDK and JoeLib2 based descriptors.
Feature Selection:
For efficient model building, selection of a preferred set of molecular descriptors is an important step. Initially, the descriptors were selected using remove uselss algorithms in weka which will remove all the descriptros that dont vary in 99% of the dataset. For further selection of relevant molecular descriptors CfSubSetEval module with best fit algorithm implemented in weka was used. Finally, an F-step approch is used to select highly relevent descriptors.
QSAR Models:
Weka is open source software widely used in the area of chemi-informatics. It is a collection of machine-learning and data-mining algorithms and supports several standard features like classification, regression, data preprocessing, and feature selection. In this work, we have used SMOreg (Sequential Minimization Optimization) implemented in Weka to predict pEC50 value of EthR compounds.
Performance Measures:
Once a regression model was constructed, goodness about the fit and statistical significance was assessed using the statistical parameters outlined below.
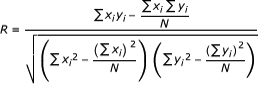
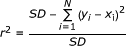 [xi and yi represent actual and predicted pEC50 value for the ith compound, N is number of compounds. SD is the sum of the Squared Deviations between the activities of the test set and mean activities of the training molecules.]
[xi and yi represent actual and predicted pEC50 value for the ith compound, N is number of compounds. SD is the sum of the Squared Deviations between the activities of the test set and mean activities of the training molecules.]
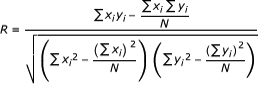
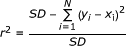 [xi and yi represent actual and predicted pEC50 value for the ith compound, N is number of compounds. SD is the sum of the Squared Deviations between the activities of the test set and mean activities of the training molecules.]
[xi and yi represent actual and predicted pEC50 value for the ith compound, N is number of compounds. SD is the sum of the Squared Deviations between the activities of the test set and mean activities of the training molecules.]