For the development of effective QSAR model, we have used total 49 molecules, which were collected from literature with their IC50 values (logarithm of 50% growth inhibitory concentration).The performance of model was evaluvated using five-fold cross-validation (Data available in supplementary dataset). Descriptor calculation:
For deriving the structural activity relationship of each molecule, we calculated descriptors using different softwares, V-life,CDK. V-life MDS (Molecular Design Suite) is a workbench for computer aided drug design (CADD) and molecule discovery. Through V-life software, we have calculated ~1002 descriptors including 1D,2D and 3D descriptors. Chemistry Development Kit (CDK), a Java based open source library for structural chemo- and bioinformatics projects calculates 178 descriptors. Descriptor calculation:
For deriving the structural activity relationship of each molecule, we calculated descriptors using different softwares, V-life,CDK. V-life MDS (Molecular Design Suite) is a workbench for computer aided drug design (CADD) and molecule discovery. Through V-life software, we have calculated ~1002 descriptors including 1D,2D and 3D descriptors. Chemistry Development Kit (CDK), a Java based open source library for structural chemo- and bioinformatics projects calculates 178 descriptors. Descriptor's selection:
In a QSAR study, selection of a preferred set of molecular descriptors is an important step to successfully derive a predictive QSAR model. Initially, the descriptors were selected using CfsubsetEval module with best fit algorithm implemented in weka. For further selection of relevant molecular descriptors F-steeping approach has been employed to remove non-significant descriptor's for the prediction of inhibitory activity against MurA enzyme. Performance Measures:
Once a regression model was constructed, goodness about the fit and statistical significance was assessed using the statistical parameters outlined below.
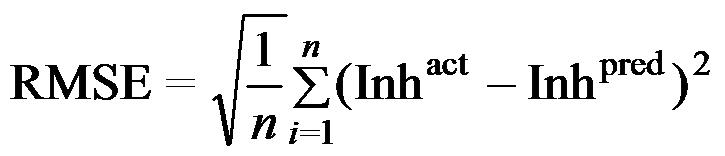
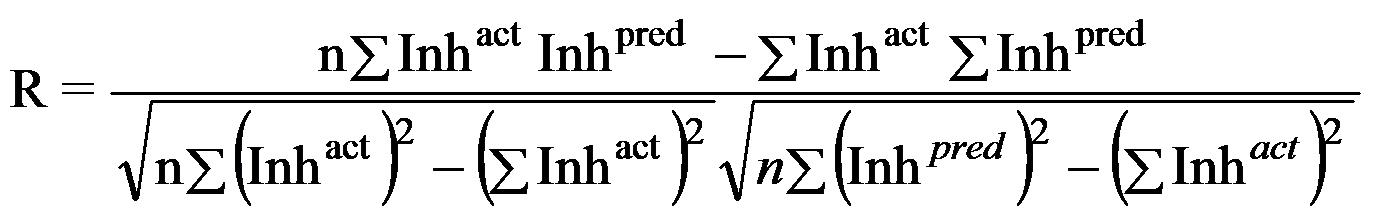
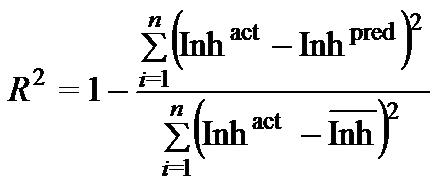
Where n is the size of test set, m is the size of training set, Toxpred is the predicted pIGC50 and Toxact is the actual pIGC50, is the toxicity in test set, RMSE are the root mean squared error, R is the Pearson's correlation coefficient between actual and predicted value, R2 (Coefficient of determination) is the statistical parameter for proportion of variability in model. The coefficient of determination is also the arithmetic average of all five folds.