

Reference: Chaudhary et., al., (2016) A Web Server and Mobile App for Computing Hemolytic Potency of Peptides. Sci Rep. 2016.
Download JAVA Standalone
1). Download the zip file HemoPI.zip from above given link. 2). After download, save it in desired location in your computer. 3). Navigate to dist folder. 4). Double click on the HemoPI.jar file or run it by using "java -jar HemoPI.jar". The software is tested on java 1.8 5). User have to select the parameters as shown below in the snapshots. 1) Home Page of HemoPI Java Standalone  2) Generating Analogs for the given Input Sequence. 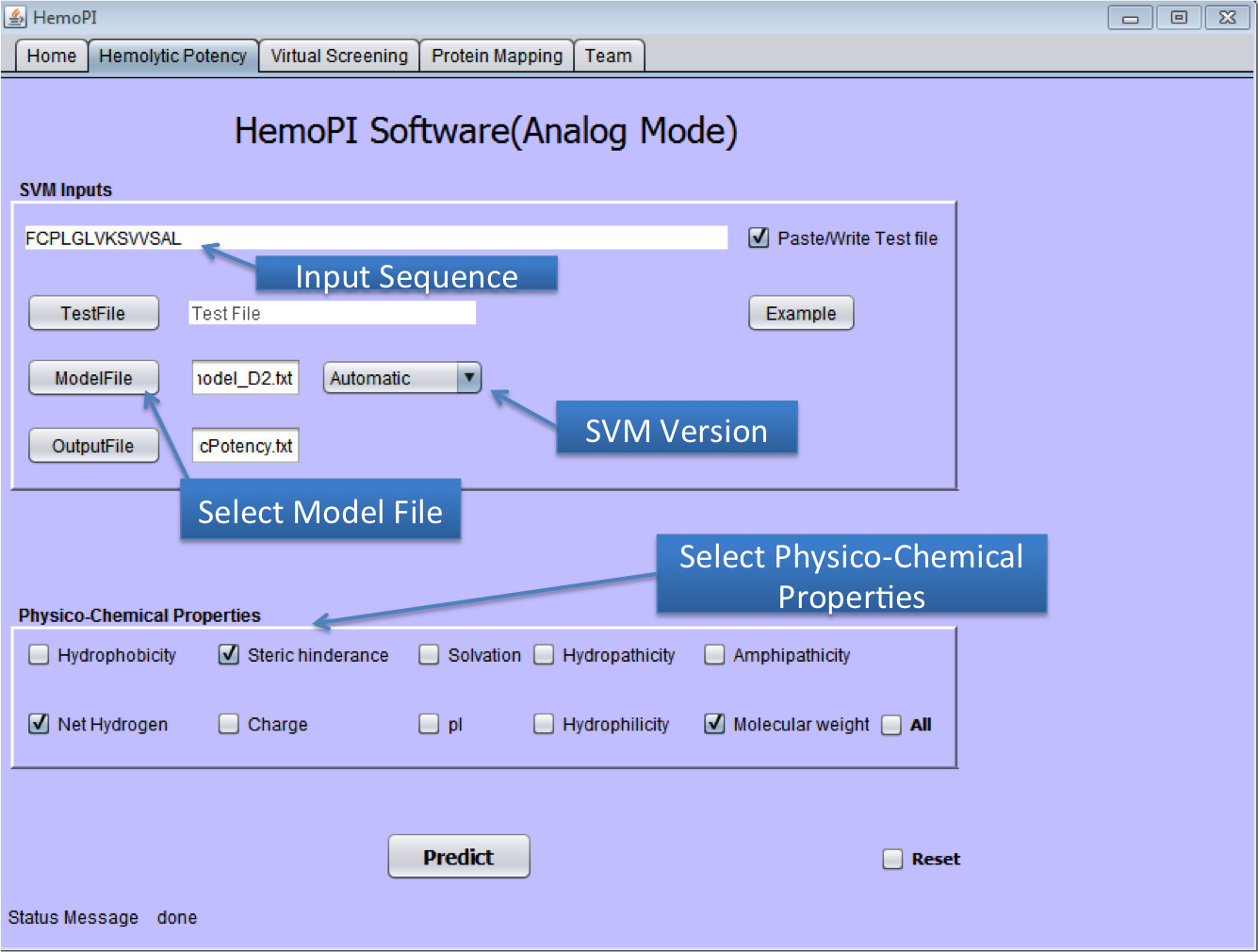 3) Result Page of Analog Tool. 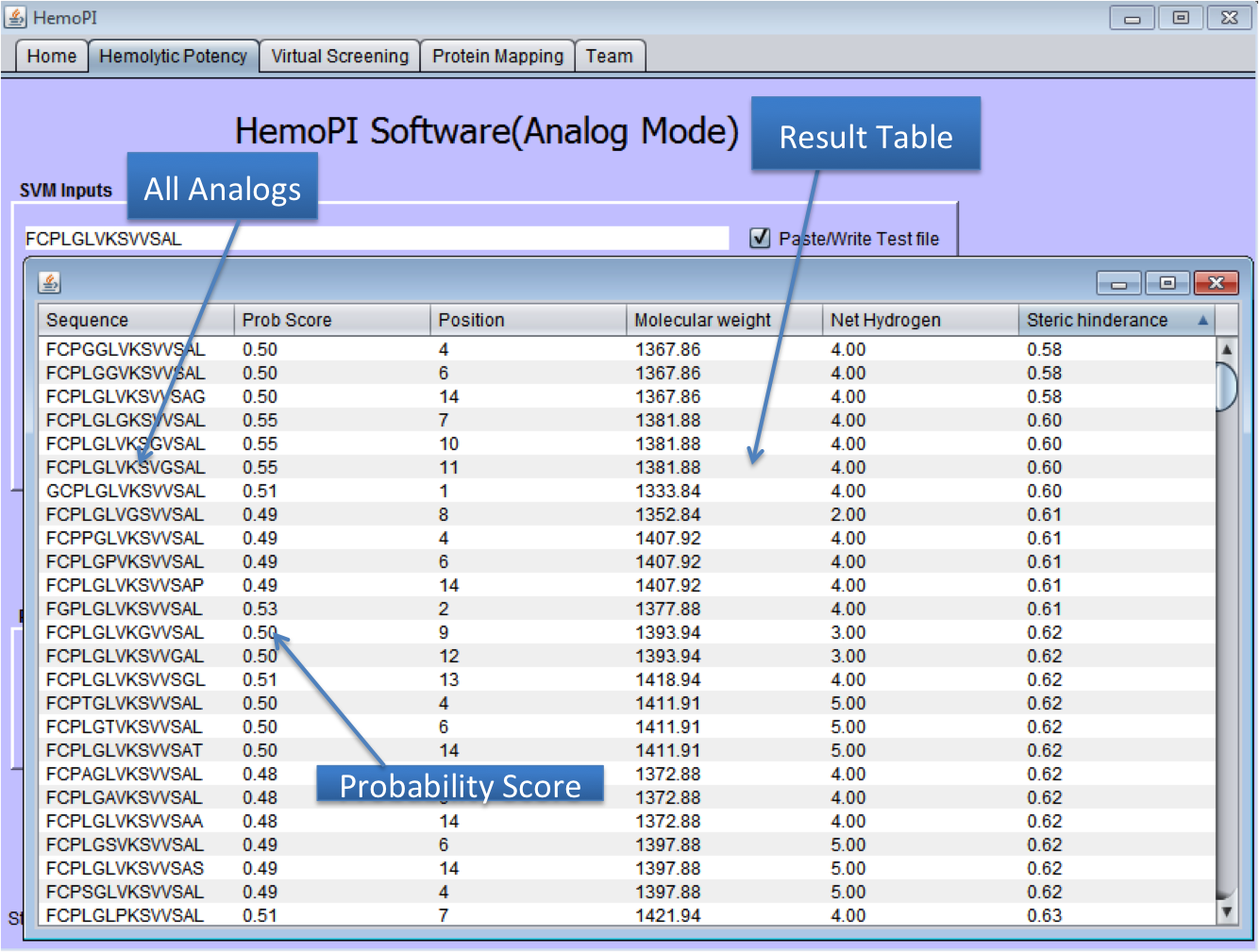 4) Virtual Screening tool(Batch Mode) of Java HemoPI.  5) Result Page of Virtual Screening Tool(Batch Mode). 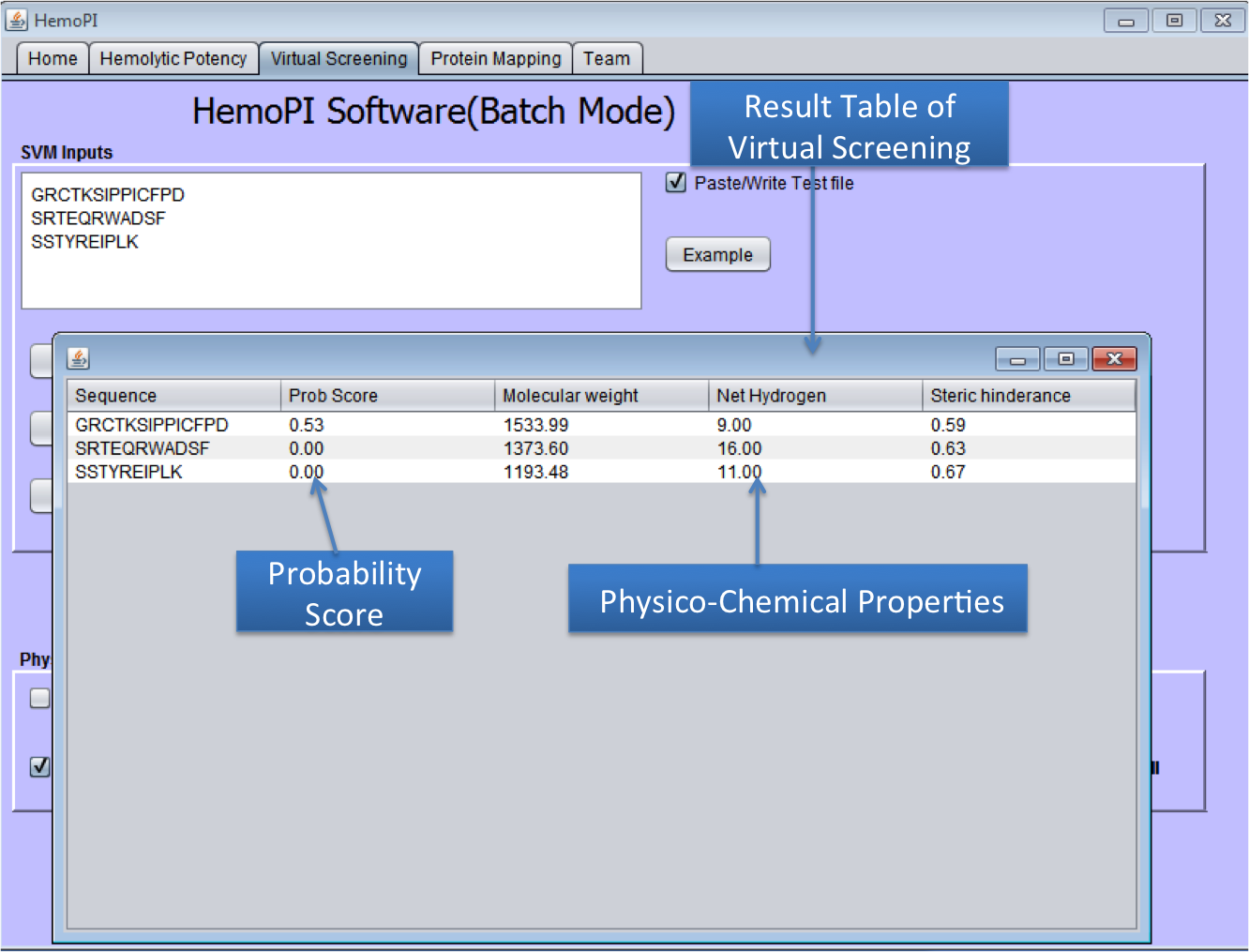 6) Protein Scan tool of Java HemoPI. 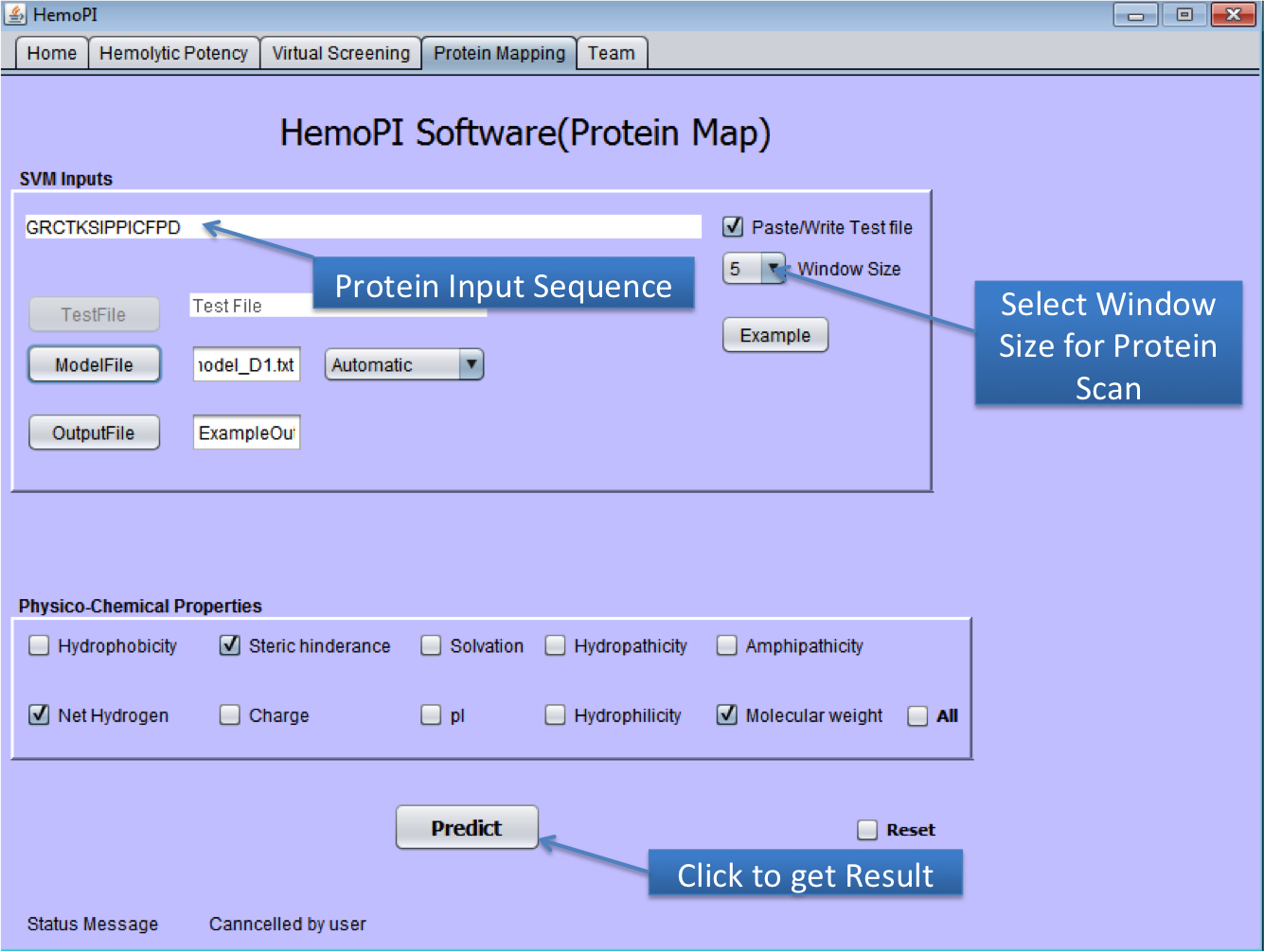 7) Result page of Protein Scan. 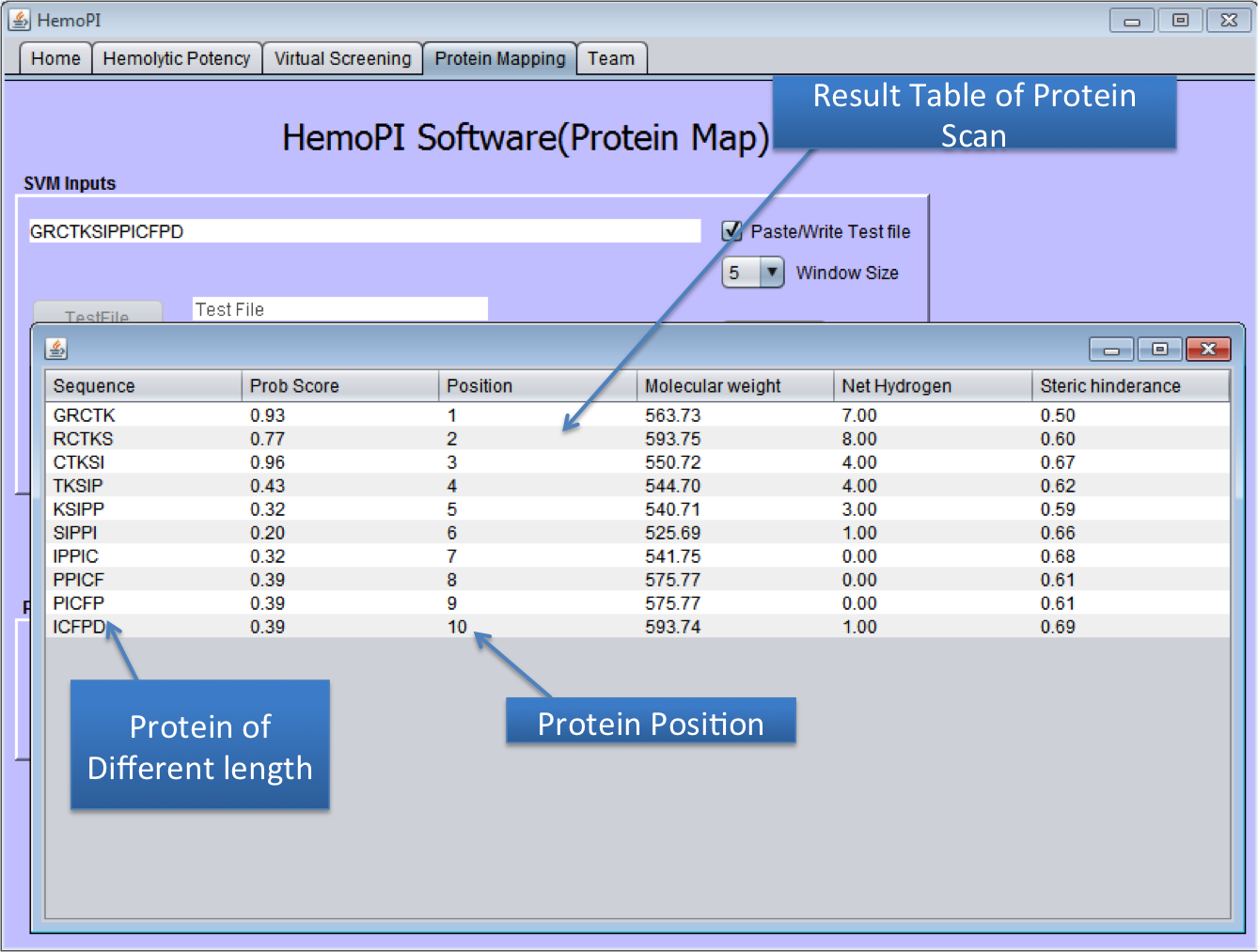 |