

Reference: Chaudhary et., al., (2016) A Web Server and Mobile App for Computing Hemolytic Potency of Peptides. Sci Rep. 2016.
Applications of HemoPI | |
|
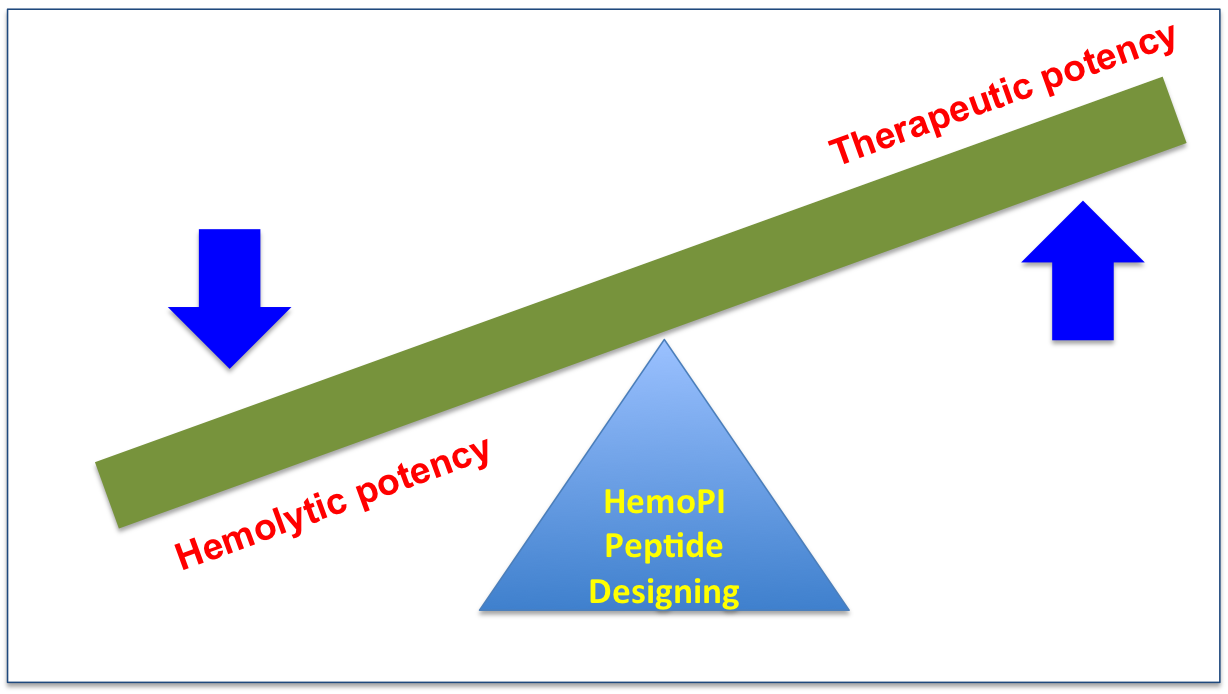
An ideal therapeutic peptide should be non-hemolytic. Many potential therapeutic peptides could not reach in clinical settings due to their high hemolytic properties. Therefore considerable efforts have been done to decrease the hemolytic properties of these peptides without compromising their therapeutic properties as shown in following figure. In this direction, HemoPI will also be useful to predict as well as design better therapeutic peptides analogs with desirable hemolytic properties.
|
|
| We have integrated various modules, which assist users to predict hemolytic properties of their peptides and also design better peptide analogs with desirable hemolytic properties. The description of these tools is as follows: |
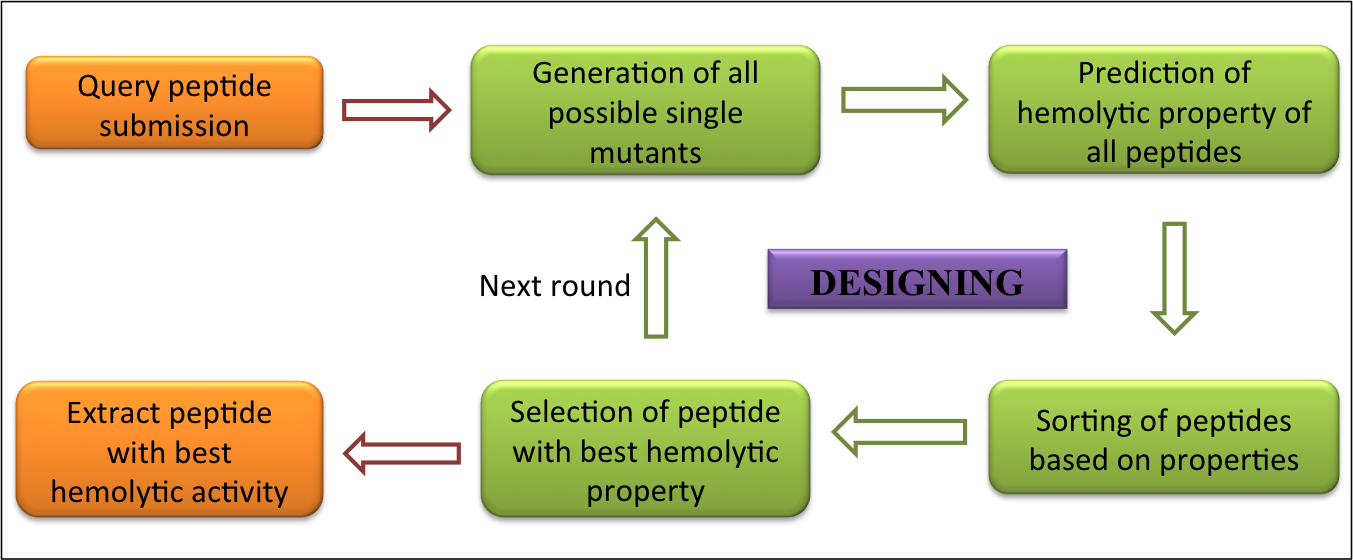
| 1. Identify: This tool predicts hemolytic or non-hemolytic property of peptide submitted by the user based on the model chosen by the user. In addition, server will also display various physicochemical properties like charge, hydrophobicity etc. This module assists users to increase/decrease hemolytic property of peptide. This tool allows the user to submit a peptide sequence and generates all possible single mutant analogs of their peptides and predicts whether the analog is hemolytic or not. For details, please visit link
|
|
| 2. Batch Submission: This tool facilitates the users to predict hemolytic properties of a large number of peptides at a time. For details, please visit link.
For details, please visit link
|
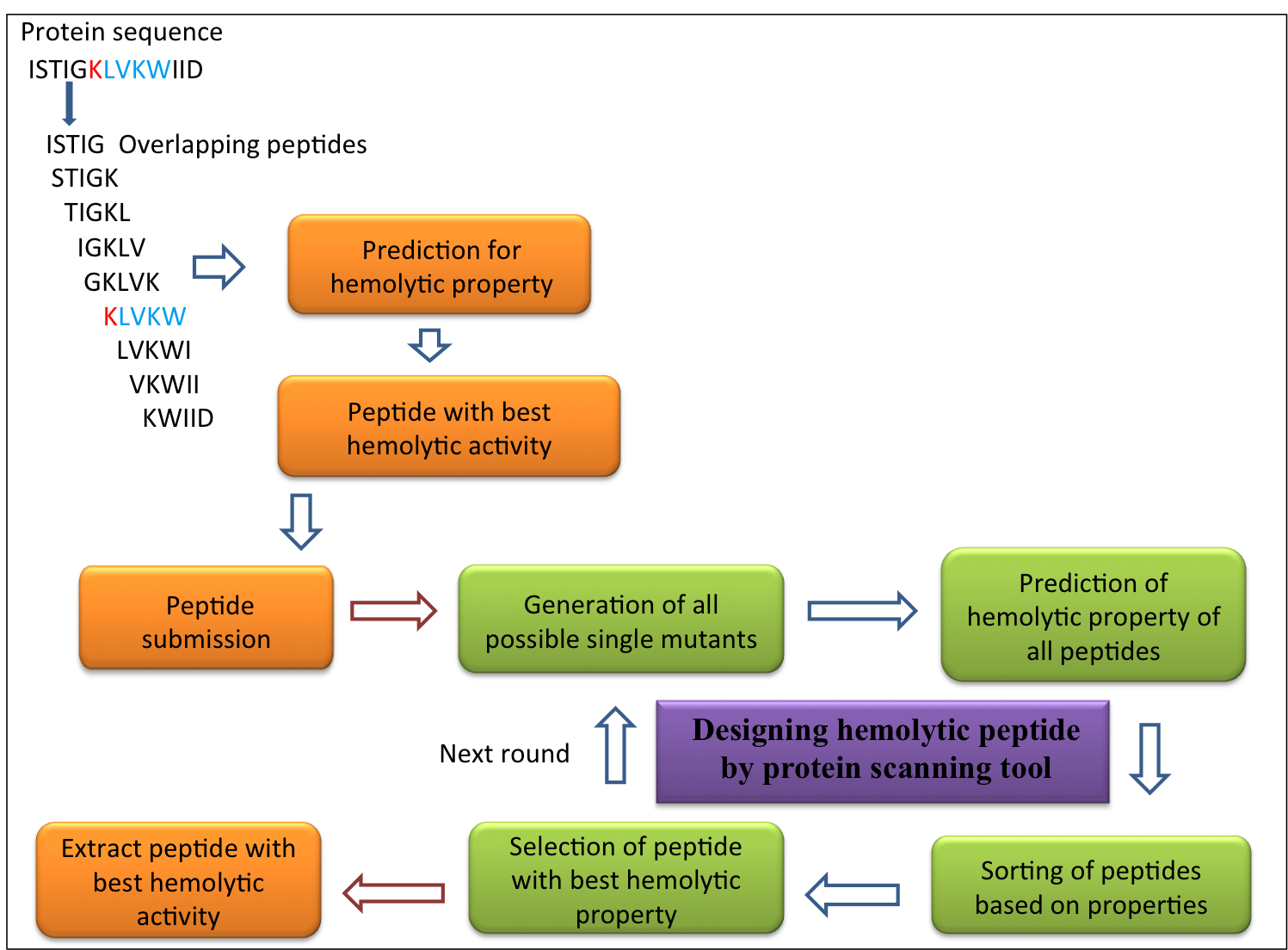
| 3. ProtScan: This module helps users to scan for the region potentially harboring hemolytic regions in their protein sequence. It allows user to select desired peptide length and then generates all possible overlapping peptides based on selected model of prediction and displayed along with their SVM score in a tabular form. For details, please visit link |
|
| 4. QMS Calculator: This module aids user in designing peptides using Quantitative Matrix based position specific score. User can obtain quantitative matrix score of each residue. Further maximum and minimum residue for that particular position along with quantitative matrix score is given. User has been provided with the option to mutate the residue to change the hemolytic potential of the peptide. For details, please visit link
|