| |||
- Aims & Objectives
- Possible Applications
- Procedure for Curation
- Architecture of CancerDR
- Database statistics
- Submission & Updates
- Data Management & system
- Requirement for access
- Disclaimer & Limitation of liability
- Copyrights
Aims & Objectives
One of the major reasons for the failure of anti-cancer therapy is the development of resistance to existing cancer drugs. There are many ways by which cancer cell develop resistance. One such reason is genetic changes in cancer causing genes leading to target alterations. CancerDR tried to correlate the target alterations with the resistance of cancer on the basis of pharmacological drug profiling on cancer cell lines.
Possible Applications
CancerDR is an ideal platform for cancer therapeutics. It has following applications.
CancerDR provides information of drug sensitivity of 148 anti-cancer drugs on 952 cancer cell lines. Due to genetic alterations in cancer causing genes, drugs showing varying sensitivity on different cell lines. CancerDR provides a platform where user can easily identify which drug class is more effective for which tissue type. This data can be extrapolated to real life after further validation.
CancerDR also provides information of all relevant targets of these drugs and their gene sequences. In addition, mutations in these genes have also been provided. This information is very helpful for user to understand the drug sensitivity/resistance in particular cancer.
CancerDR provide structures of all wild type and their mutated drug targets. A tool has also been provided for user, where mutated and wild type target structures can be compared to understand the possible changes occurred in the mutated target.
Sequencing cost is decreasing day by day, so it is now possible to sequence the whole genome of cancer patients. An important tool has been integrated in CancerDR, where user can submit their short reads and can map these reads to all cancer causing genes. By doing so, user can identify all possible mutations in their sequence of interest.
By analyzing genetic alterations in drug targets and drug sensitivity profiles of drugs user can design or select the best therapeutic options for a particular cancer.
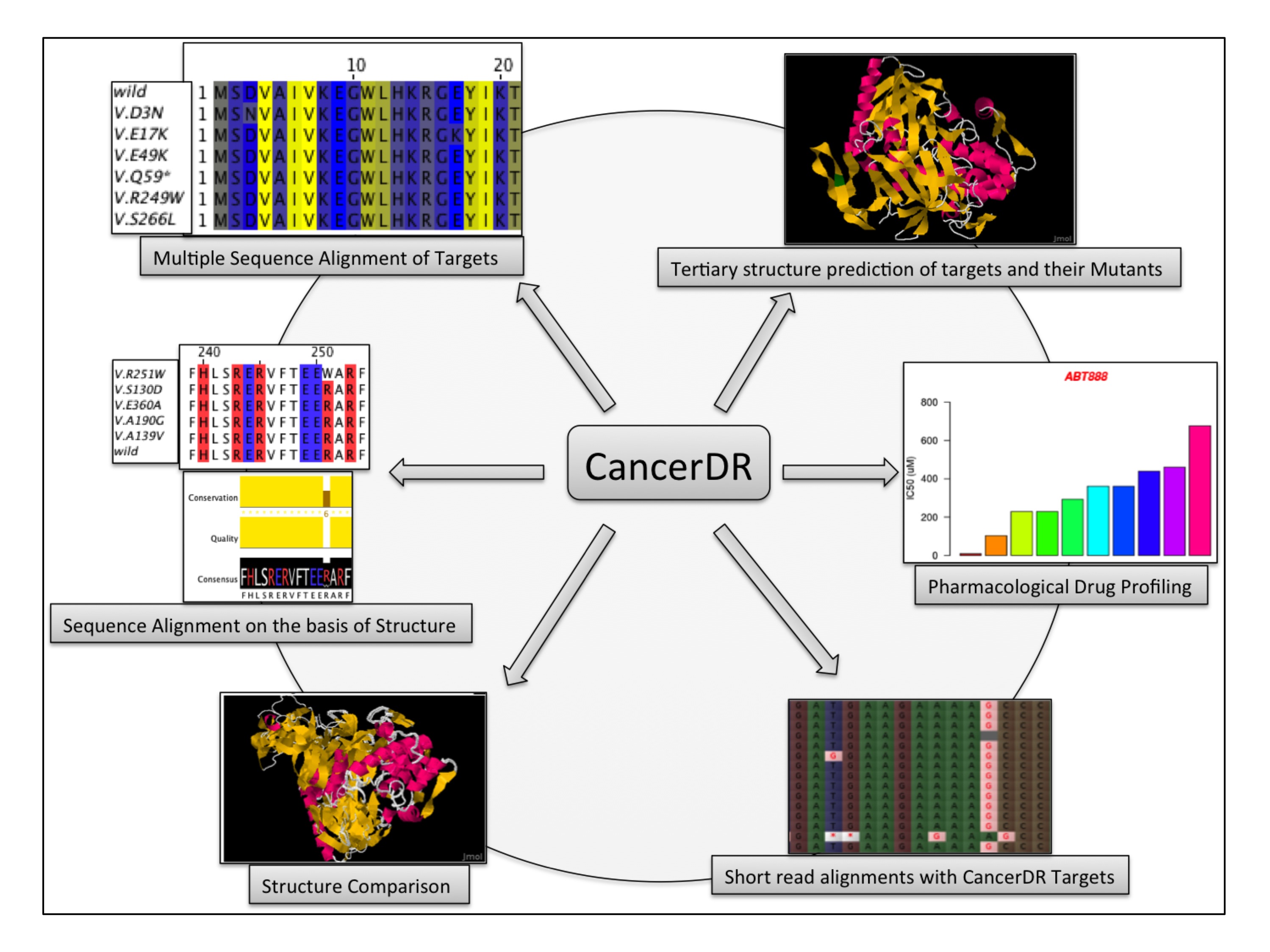
Procedure for Curation
Aim of the CancerDR is to collect and compile the pharmacological profiling of anticancer drugs on a large number of cancer cell lines in relation to the mutation status of the drug target genes. For this, we collected the pharmacological profiling data of 148 anticancer drugs across 952 cancer cell lines from two sources, COSMIC and CCLE. In release 2 of COSMIC, 138 anticancer drugs targeting a wide range of therapeutic targets were screened on 714 cancer cell lines and in case of CCLE, 24 drugs were screened on 503 cancer cell lines. We focused on 116 drug targets and their mutation status in each cancer cell line, which was collected from the hybrid capture sequencing data of 947 cancer cell lines available on CCLE website. Mutations reported in sequencing data of CCLE for 116 drug targets were mapped on their sequences by Perl programming and these mutated sequences are available for download at CancerDR website.
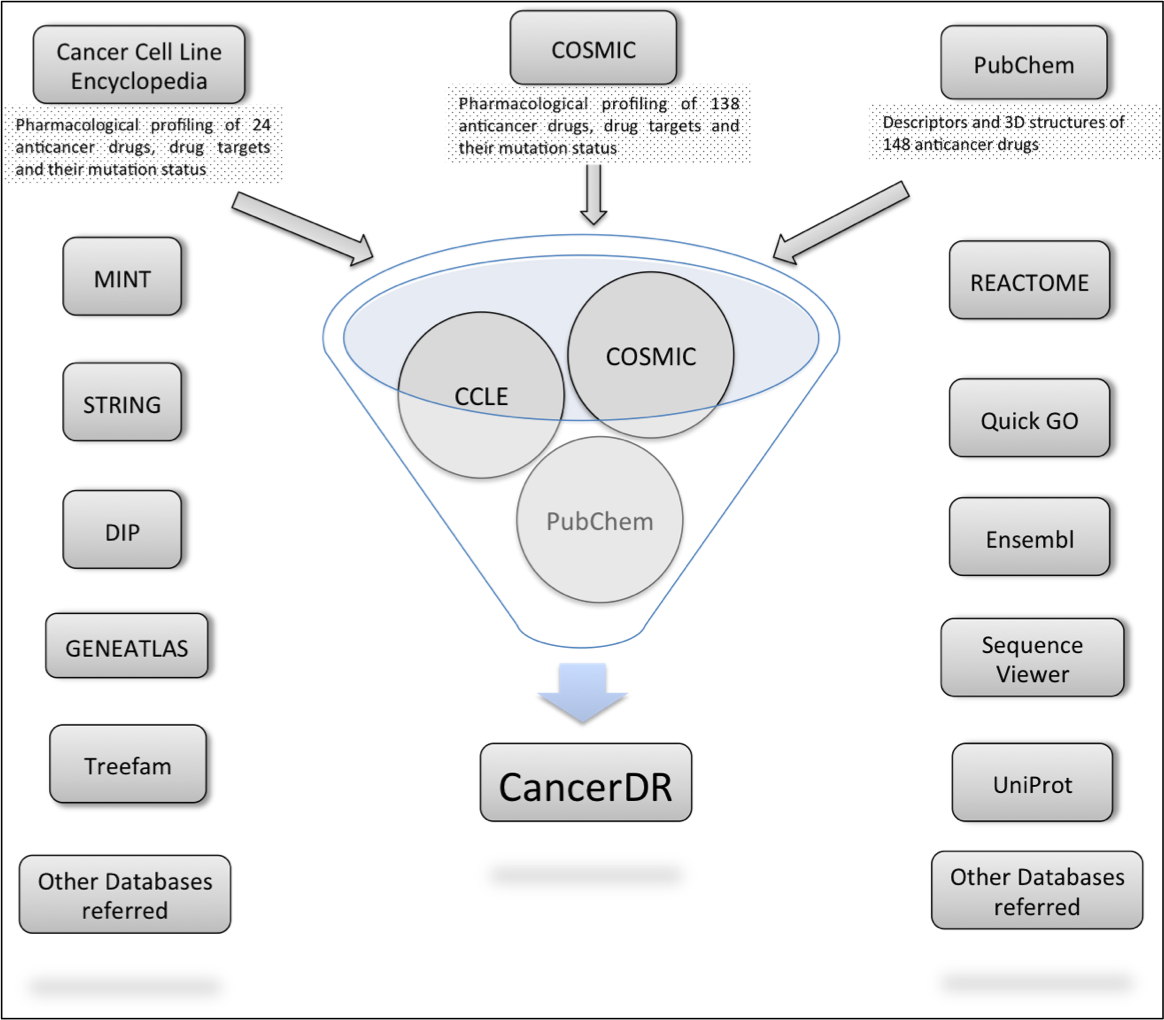
Architecture of CancerDR
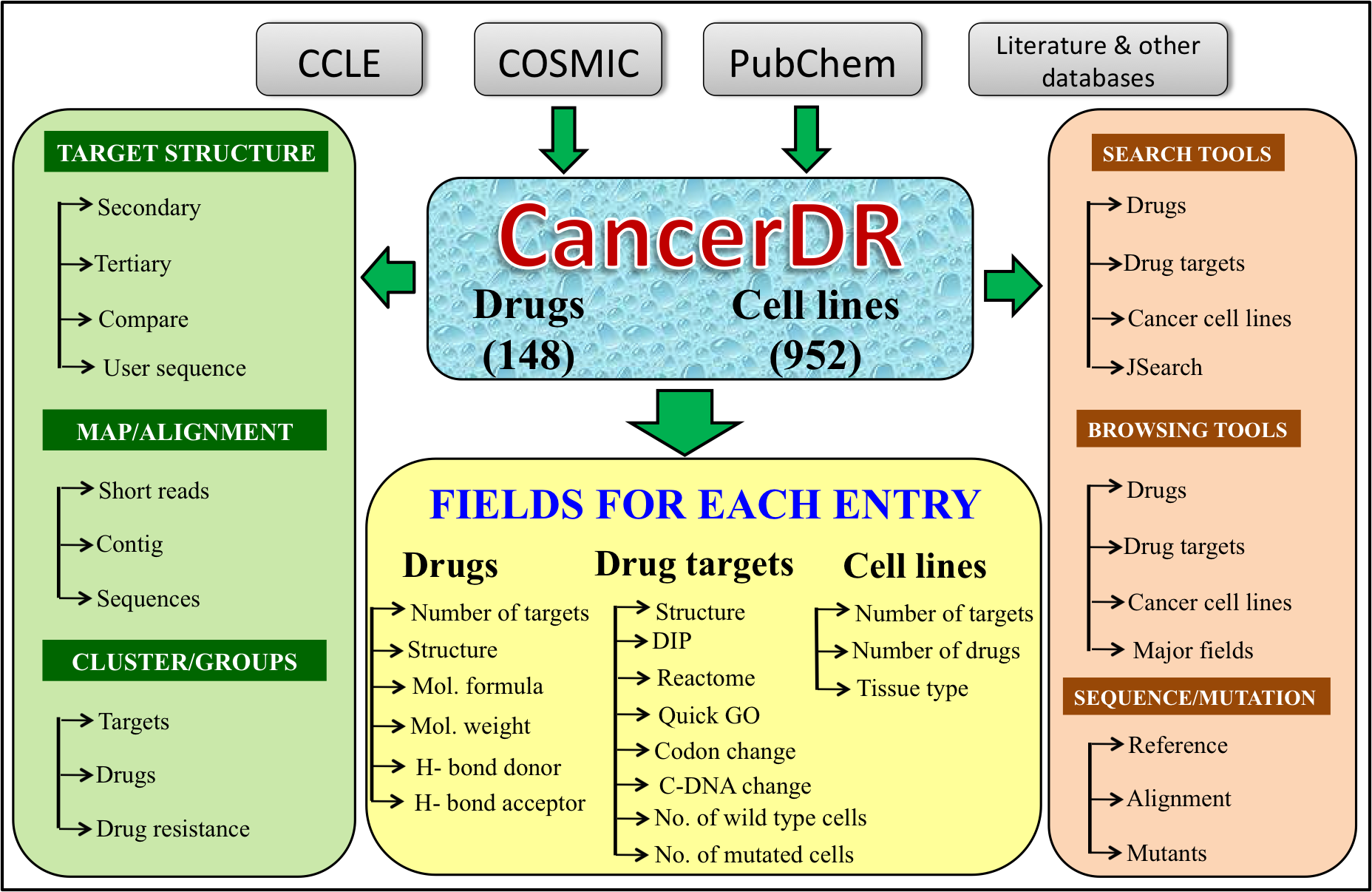
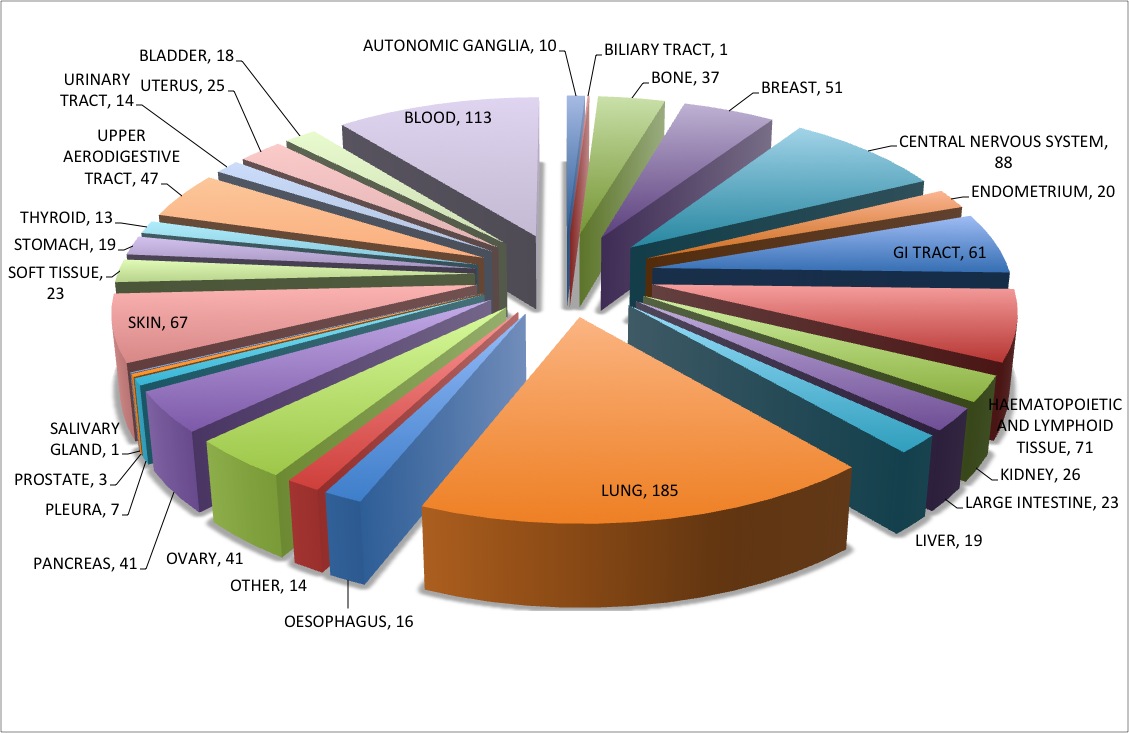
Database statistics
We have covered around 148 anticancer drugs assayed in CCLE and COSMIC, which are targeting a wide range of therapeutic cancer targets.
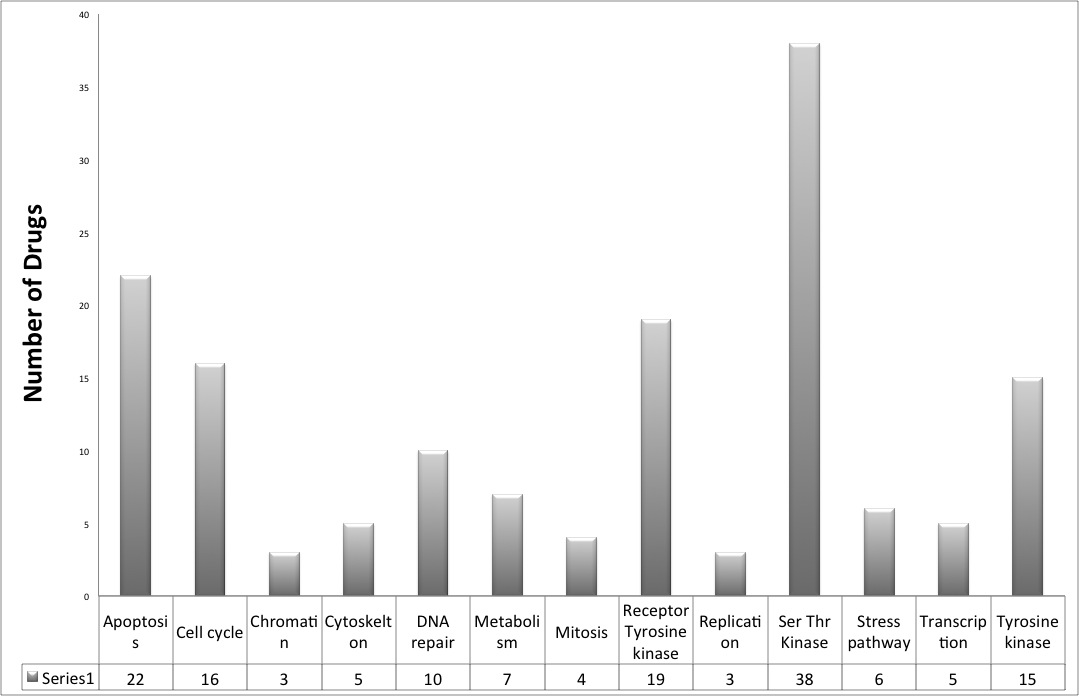
952 cell lines from different tissue origins are covered in CancerDR.
Clustering in CancerDR
Clustering of cell lines and drugs, cluster the cell line or drug according to the range of IC50 (µM). Two kind of ranges are used in CancerDR. First, in which ranges are made in multiples of sensitivity reference. Sensitivity reference is the lowest IC50 value reported for particular drug or cell line. In second type of clustering, absolute range is used i.e. R1: 0-0.001µM, R2: 0.001-0.005µM, R3: 0.005-0.025µM, R4: 0.025-0.125µM, R5: 0.125-0.625µM, R6: 0.625-15µM, R7: 15-390µM, R8: greater than 390µM. Clustering can be done either according to the tissue types in which cells of particular tissue type will cluster or cell lines having some mutations in drug target. Clustering is very useful in understanding, which drug is most effective to what kind of tissue types and at the same time, it also tells about which cell line or tissue type is most sensitive or resistant to which drug.
Submission & Updates
The CancerDR web server allows users to submit their data online by filling out a HTML form. Before inclusion in CancerDR we will confirm the validity of the supplied data, in order to maintain quality. We continue to search the literature and will add more drug their targets and cell lines. In order to maintain the consistency we will release database time to time. All entries thus obtained will be available as new submissions. The success of this database is highly dependent upon contributors: users are requested to submit data.
Data Management & system
CancerDR has been three main tables named drug_table, target_table and cell_table. Drug_table contain detail information about each drug; target_table contains information about target of drugs particularly sequence; cell_table provides information about various types of cancer cell lines. All tables are maintained in Relational Database Management System? (RDBMS) called MySQL which is a public domain software freely available to public.
Structures of cancer drug targets are stored in Protein DataBank (PDB) format. All structures are downloadable from our web site. In order to provide updates and easy download we are providing rsync facility.
Requirement for access
CancerDR web server is developed in a Linux environment on Red Hat Linux. This server is designed to provide easy access to the user. Methods for searching the databases and displaying the selected objects were built with a combination of Java Scripts, PHP and PERL.
System requirements for running the CancerDR System: It may be run on any modern browser, we have checked on Firefox, Google Chrome. Some page may require java plugin, particularly for viewing structure (Jmol) and alignment (Jalview).
Disclaimer & Limitation of liability
Data, information, e-mail addresses, hyperlinks (URLs) and data analysis tools provided by BIC, IMTECH New Delhi are on an "as is" basis, without any warranties. IMTECH and the developers of CancerDR give no warranty, including without limitation to the quality, accuracy or completeness of data or information, or that the use of data or information or data analysis tools will not infringe any patent, or intellectual property rights of any party. IMTECH shall not be liable for any claim, including without limitation, for any kind of loss, harm, illness, damage, injury originating from access to or use of data, information, e-mail addresses, hyperlinks (URLs) and data analysis tools, even if IMTECH advises of the possibility of such damages.
Copyrights
Copyright 2012; CancerDR: Cancer Drug Resistance Database