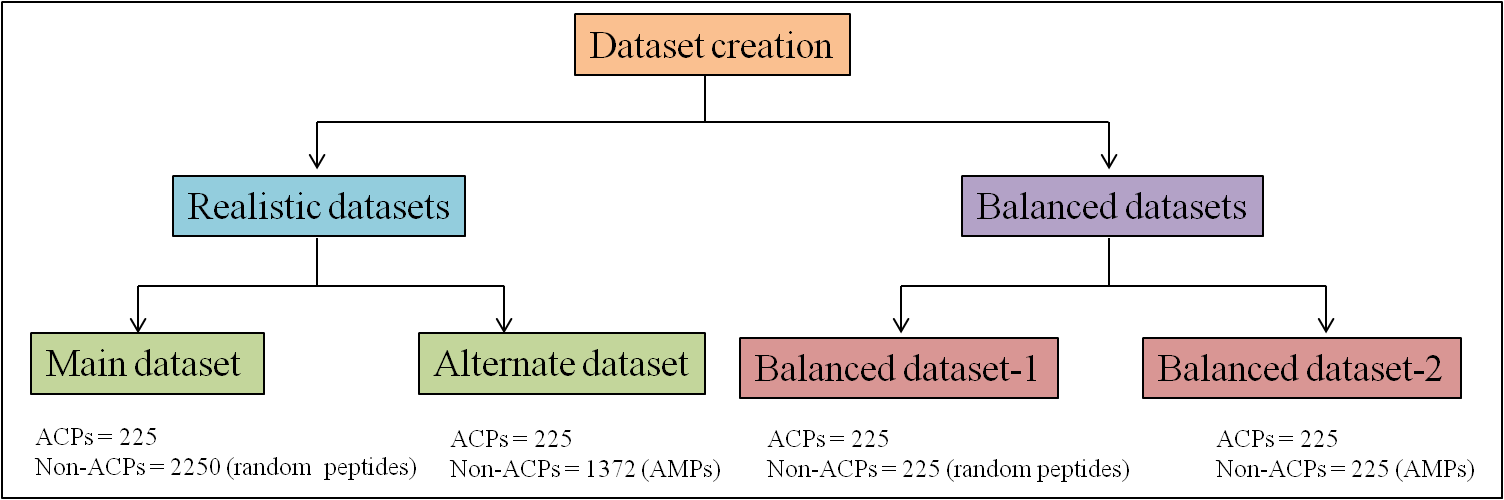
Main dataset
We have extracted 225 experimentally validated ACPs from literature and databases like antimicrobial database (ADP), collection of antimicrobial peptides (CAMP) and database of anuran defense peptides (DADP). Majority of these peptides are AMPs with anticancer activities. All these peptides were unique and considered as positive examples. Since there are very few experimentally proved non-ACPs, we extracted (2250) random peptides from SwissProt proteins and considered them as negative examples (non-ACP). Thus, main dataset contains 225 ACPs and 2250 non-ACPs.
Alternate dataset
In this dataset, we have considered 225 experimentally validated ACPs as postive examples. We have extracted 1372 AMPs to which no anticancer activity were reported, from APD database and considered as negative examples (non-ACPs). Thus, this datset contains 225 ACPs and 1372 non-ACPs. This dataset was used to develop SVM models, which discriminate ACPs from AMPs.
Balanced datasets
In order to remove the biasness in machine learning, we have created balanced datasets from these realistic datasets.
Balanced dataset-1
We have extrated 225 random peptides from 2250 Swissprot peptides to creat a balanced dataset.
Balanced dataset-2
It contains 225 experimentally validated ACPs as positive exmples as stated above and equal number of AMPs were selected from 1372 AMPs extracted from APD database.